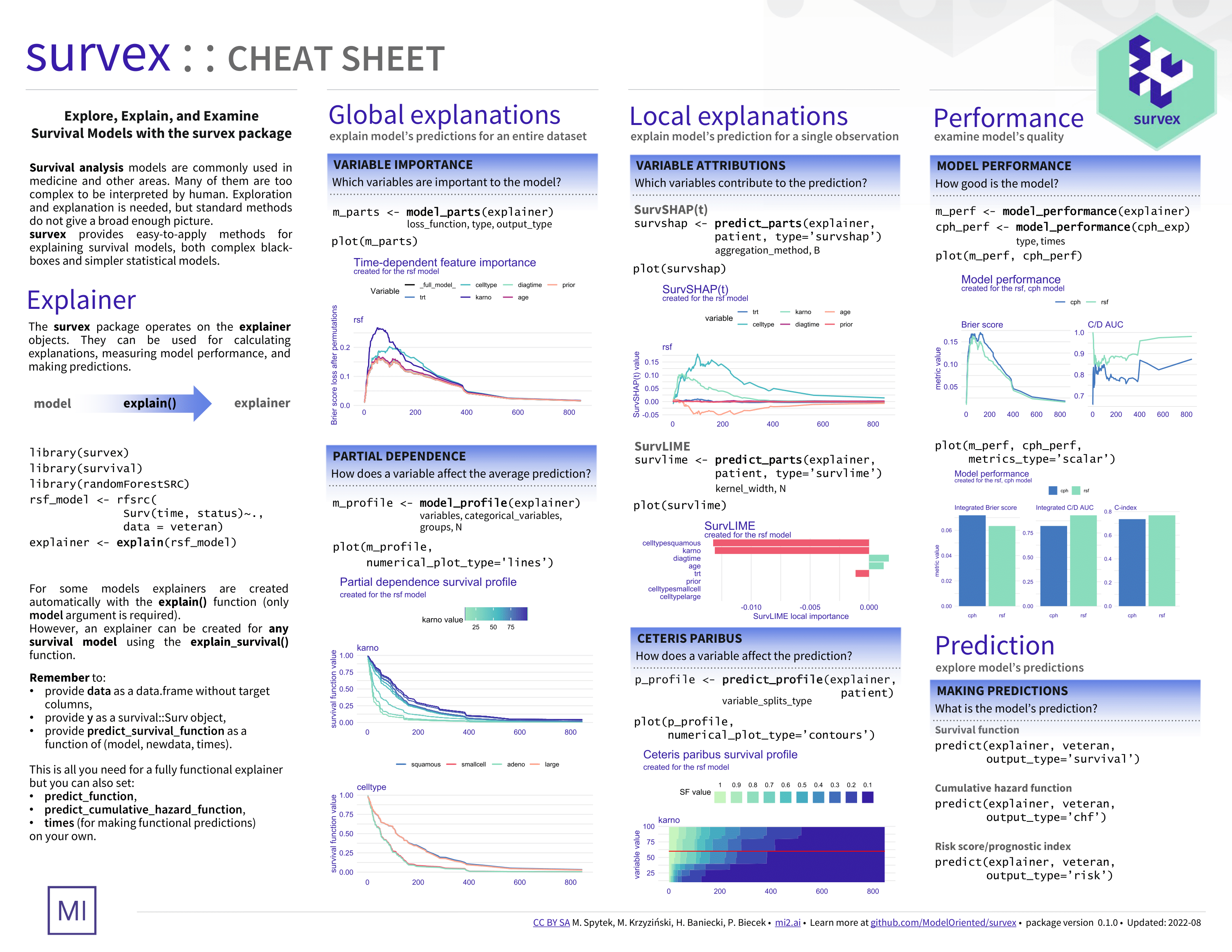
Overview
Survival analysis is a task dealing with time-to-event prediction. Aside from the well-understood models like CPH, many more complex models have recently emerged, but most lack interpretability. Due to a functional type of prediction, either in the form of a survival function or a cumulative hazard function, standard model-agnostic explanations cannot be applied directly.
The survex package provides model-agnostic explanations for machine learning survival models. It is based on the DALEX package. If you’re unfamiliar with explainable machine learning, consider referring to the Explanatory Model Analysis book – most of the methods included in survex extend these described in EMA and implemented in DALEX but to models with functional output.
The main explain() function uses a model and data to create a standardized explainer object, which is further used as an interface for calculating predictions. We automate creating explainers from the following packages: mlr3proba, censored, ranger, randomForestSRC, and survival. Raise an Issue on GitHub if you find models from other packages that we can incorporate into the explain() interface.
Note that an explainer can be created for any survival model, using the explain_survival() function by passing model, data, y, and predict_survival_function arguments.
Installation
The package is available on CRAN:
install.packages("survex")The latest development version can be installed from GitHub using devtools::install_github():
devtools::install_github("https://github.com/ModelOriented/survex")Simple demo
library("survex")
library("survival")
library("ranger")
# create a model
model <- ranger(Surv(time, status) ~ ., data = veteran)
# create an explainer
explainer <- explain(model,
data = veteran[, -c(3, 4)],
y = Surv(veteran$time, veteran$status))
# evaluate the model
model_performance(explainer)
# visualize permutation-based feature importance
plot(model_parts(explainer))
# explain one prediction with SurvSHAP(t)
plot(predict_parts(explainer, veteran[1, -c(3, 4)]))Functionalities and roadmap
Existing functionalities: - [x] unified prediction interface using the explainer object - predict() - [x] calculation of performance metrics (Brier Score, Time-dependent C/D AUC, metrics from mlr3proba) - model_performance() - [x] calculation of feature importance (Permutation Feature Importance - PFI) - model_parts() - [x] calculation of partial dependence (Partial Dependence Profiles - PDP, Accumulated Local Effects - ALE) - model_profile() - [x] calculation of 2-dimensional partial dependence (2D PDP, 2D ALE) - model_profile_2d() - [x] calculation of local feature attributions (SurvSHAP(t), SurvLIME) - predict_parts() - [x] calculation of local ceteris paribus explanations (Ceteris Paribus profiles - CP/ Individual Conditional Expectations - ICE) - predict_profile() - [x] calculation of global feature attributions using SurvSHAP(t) - model_survshap()
Currently in develompment: - [ ] …
Future plans: - [ ] … (raise an Issue on GitHub if you have any suggestions) - [ ] examples for sursvm and survboost (https://github.com/ModelOriented/survex/issues/88)
Citation
If you use survex, please cite our article:
M. Spytek, M. Krzyziński, S. H. Langbein, H. Baniecki, M. N. Wright, P. Biecek. survex: an R package for explaining machine learning survival models. Bioinformatics, Volume 39, Issue 12, btad723, 2023.
@article{spytek2023survex,
author = {Mikołaj Spytek and Mateusz Krzyziński and Sophie Hanna Langbein and
Hubert Baniecki and Marvin N Wright and Przemysław Biecek},
title = {survex: an {R} package for explaining machine learning survival models},
journal = {Bioinformatics},
volume = {39},
number = {12},
pages = {btad723},
year = {2023},
month = {12},
doi = {10.1093/bioinformatics/btad723}
}Applications of survex
- H. Baniecki, B. Sobieski, P. Bombiński, P. Szatkowski, P. Biecek. Hospital Length of Stay Prediction Based on Multi-modal Data towards Trustworthy Human-AI Collaboration in Radiomics. International Conference on Artificial Intelligence in Medicine, 2023.
- W. Chen, B. Zhou, C. Y. Jeon, F. Xie, Y.-C. Lin, R. K. Butler, Y. Zhou, T. Q. Luong, E. Lustigova, J. R. Pisegna, B. U. Wu. Machine learning versus regression for prediction of sporadic pancreatic cancer. Pancreatology, 2023.
- M. Nachit, Y. Horsmans, R. M. Summers, I. A. Leclercq, P. J. Pickhardt. AI-based CT Body Composition Identifies Myosteatosis as Key Mortality Predictor in Asymptomatic Adults. Radiology, 2023.
- R. Passera, S. Zompi, J. Gill, A. Busca. Explainable Machine Learning (XAI) for Survival in Bone Marrow Transplantation Trials: A Technical Report. BioMedInformatics, 2023.
- P. Donizy, M. Spytek, M. Krzyziński, K. Kotowski, A. Markiewicz, B. Romanowska-Dixon, P. Biecek, M. P. Hoang. Ki67 is a better marker than PRAME in risk stratification of BAP1-positive and BAP1-loss uveal melanomas. British Journal of Ophthalmology, 2023.
- X. Qi, Y. Ge, A. Yang, Y. Liu, Q. Wang, G. Wu. Potential value of mitochondrial regulatory pathways in the clinical application of clear cell renal cell carcinoma: a machine learning-based study. Journal of Cancer Research and Clinical Oncology, 2023.
- C. C. Lee, S. Y. Su, S. F. Sung. Machine learning-based survival analysis approaches for predicting the risk of pneumonia post-stroke discharge. International Journal of Medical Informatics, 2024.
- P. Wang, X. Qian, W. Jiang, H. Wang, Y. Wang, Y. Zhou, Y. Zhang, Y. Huang, X. Zhai. Cord Blood Transplantation for Very Early‑Onset Inflammatory Bowel Disease Caused by Interleukin‑10 Receptor Deficiency. Journal of Clinical Immunology, 2024.
- E. Ruiz, J. Honles, R. Fernández, K. Uribe, J. P. Cerapio, K. Cancino, J. Contreras-Mancilla, S. Casavilca-Zambrano, F. Berrospi, P. Pineau, S. Bertani. A preoperative risk score based on early recurrence for estimating outcomes after resection of hepatocellular carcinoma in the non-cirrhotic liver. HPB, 2024.
- Share it with us!
Related work
- H. Ishwaran, U. B. Kogalur, E. H. Blackstone, M. S. Lauer. Random survival forests. Annals of Applied Statistics, 2008.
- A. Grudziąż, A. Gosiewska, P. Biecek. survxai: an R package for structure-agnostic explanations of survival models. Journal of Open Source Software, 2018.
- M. S. Kovalev, L. V. Utkin, E. M. Kasimov. SurvLIME: A method for explaining machine learning survival models. Knowledge-Based Systems, 2020.
- R. Sonabend, F. J. Király, A. Bender, B. Bischl, M. Lang. mlr3proba: an R package for machine learning in survival analysis. Bioinformatics, 2021.
- E. Hvitfeldt, H. Frick. censored: ‘parsnip’ Engines for Survival Models. CRAN v0.1.0, 2022.
- M. Krzyziński, M. Spytek, H. Baniecki, P. Biecek. SurvSHAP(t): Time-dependent explanations of machine learning survival models. Knowledge-Based Systems, 2023.