Instance Level Parts of Survival Model Predictions
Source:R/predict_parts.R
predict_parts.surv_explainer.RdThis function decomposes the model prediction into individual parts, which are attributions of particular variables. The explanations can be made via the SurvLIME and SurvSHAP(t) methods.
predict_parts(explainer, ...)
# S3 method for surv_explainer
predict_parts(
explainer,
new_observation,
...,
N = NULL,
type = "survshap",
output_type = "survival",
explanation_label = NULL
)Arguments
- explainer
an explainer object - model preprocessed by the
explain()function- ...
other parameters which are passed to
iBreakDown::break_downifoutput_type=="risk", or ifoutput_type=="survival"tosurv_shap()orsurv_lime()functions depending on the selected type- new_observation
a new observation for which prediction need to be explained
- N
the number of observations used for calculation of attributions. If
NULL(default) all explainer data will be used for SurvSHAP(t) and 100 neigbours for SurvLIME.- type
if
output_type == "survival"must be either"survshap"or"survlime", otherwise refer to theDALEX::predict_parts- output_type
either
"survival","chf"or"risk"the type of survival model output that should be considered for explanations. If"survival"the explanations are based on the survival function. If"chf"the explanations are based on the cumulative hazard function. Otherwise the scalar risk predictions are used by theDALEX::predict_partsfunction.- explanation_label
a label that can overwrite explainer label (useful for multiple explanations for the same explainer/model)
Value
An object of class "predict_parts_survival" and additional classes depending on the type of explanations. It is a list with the element result containing the results of the calculation.
Additional parameters
There are additional parameters that are passed to internal functions
for
survlimeN- a positive integer, number of observations generated in the neighbourhooddistance_metric- character, name of the distance metric to be used, only"euclidean"is implementedkernel_width- a numeric or"silverman", parameter used for calculating weights, by default it'ssqrt(ncol(data)*0.75). If"silverman"the kernel width is calculated using the method proposed by Silverman and used in the SurvLIMEpy Python package.sampling_method- character, name of the method of generating neighbourhood, only"gaussian"is implementedsample_around_instance- logical, if the neighbourhood should be generated with the new observation as the center (default), or should the mean of the whole dataset be used as the centermax_iter- a numeric, maximal number of iteration for the optimization problemcategorical_variables- character vector, names of variables that should be treated as categories (factors are included by default)k- a small positive number > 1, added to chf before taking log, so that weigths aren't negative
for
survshapy_true- a two element numeric vector or matrix of one row and two columns, the first element being the true observed time and the second the status of the observation, used for plottingcalculation_method- a character, either"kernelshap"for use ofkernelshaplibrary (providing faster Kernel SHAP with refinements) or"exact_kernel"for exact Kernel SHAP estimationaggregation_method- a character, either"mean_absolute"or"integral","max_absolute","sum_of_squares"
References
[1] Krzyziński, Mateusz, et al. "SurvSHAP(t): Time-dependent explanations of machine learning survival models." Knowledge-Based Systems 262 (2023): 110234
[2] Kovalev, Maxim S., et al. "SurvLIME: A method for explaining machine learning survival models." Knowledge-Based Systems 203 (2020): 106164.
[3] Pachón-García, Cristian, et al. "SurvLIMEpy: A Python package implementing SurvLIME." Expert Systems with Applications 237 (2024): 121620.
Examples
# \donttest{
library(survival)
library(survex)
cph <- coxph(Surv(time, status) ~ ., data = veteran, model = TRUE, x = TRUE, y = TRUE)
cph_exp <- explain(cph)
#> Preparation of a new explainer is initiated
#> -> model label : coxph ( default )
#> -> data : 137 rows 6 cols ( extracted from the model )
#> -> target variable : 137 values ( 128 events and 9 censored , censoring rate = 0.066 ) ( extracted from the model )
#> -> times : 50 unique time points , min = 1.5 , median survival time = 80 , max = 999
#> -> times : ( generated from y as uniformly distributed survival quantiles based on Kaplan-Meier estimator )
#> -> predict function : predict.coxph with type = 'risk' will be used ( default )
#> -> predict survival function : predictSurvProb.coxph will be used ( default )
#> -> predict cumulative hazard function : -log(predict_survival_function) will be used ( default )
#> -> model_info : package survival , ver. 3.7.0 , task survival ( default )
#> A new explainer has been created!
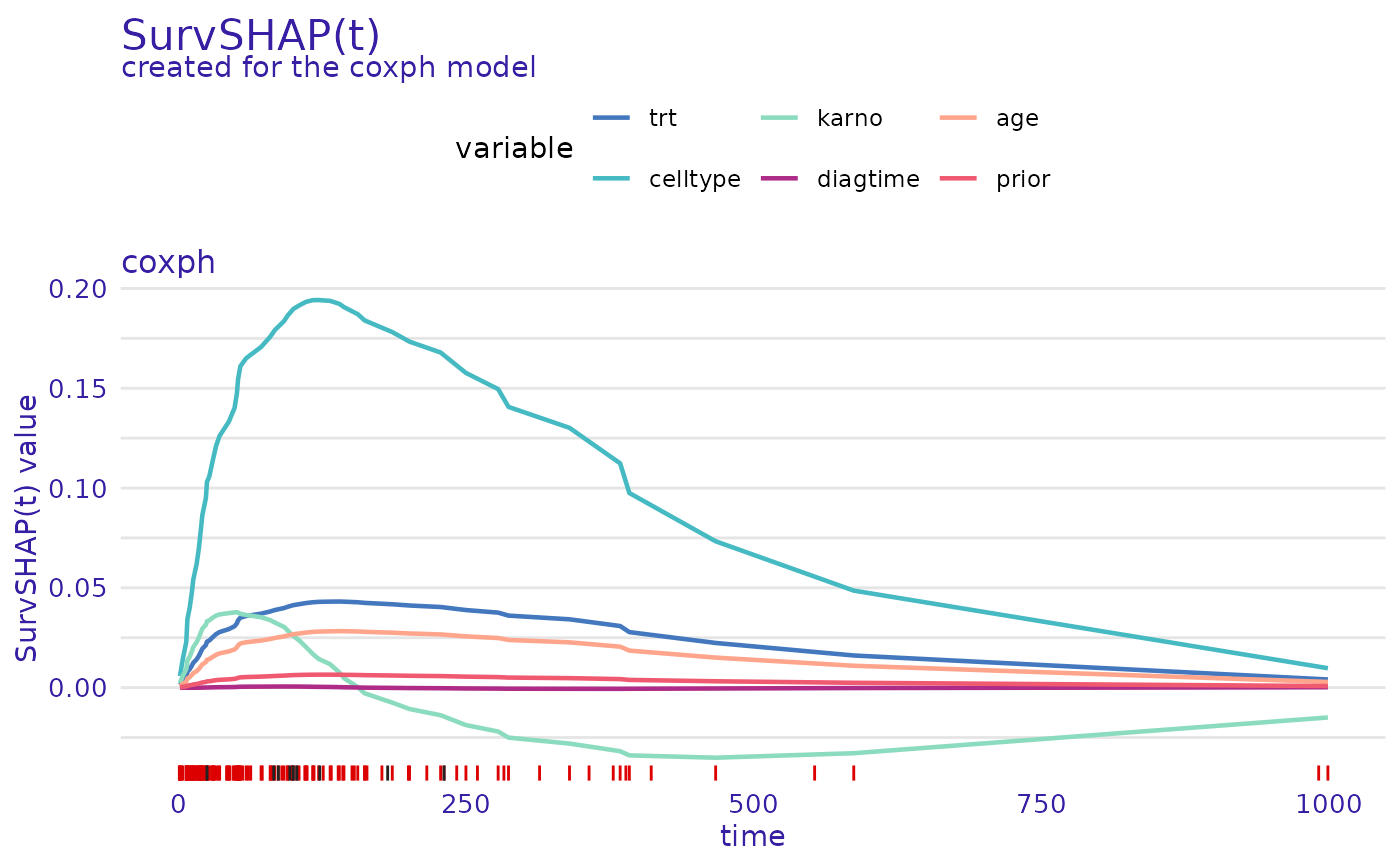
cph_predict_parts_survshap <- predict_parts(cph_exp, new_observation = veteran[1, -c(3, 4)])
head(cph_predict_parts_survshap$result)
#> trt celltype karno diagtime age
#> t=1.5 0.001358479 0.005752629 0.002400785 -1.753141e-05 0.0007519409
#> t=4 0.003364680 0.014321361 0.005877960 -3.897254e-05 0.0018795538
#> t=7 0.005360141 0.022931960 0.009247829 -5.480637e-05 0.0030211319
#> t=8 0.007989085 0.034411531 0.013535810 -6.687523e-05 0.0045549833
#> t=10 0.009277550 0.040094562 0.015567469 -6.898441e-05 0.0053188972
#> t=12 0.011153127 0.048434497 0.018433696 -6.740775e-05 0.0064449115
#> prior
#> t=1.5 0.0001596961
#> t=4 0.0004003893
#> t=7 0.0006457473
#> t=8 0.0009784720
#> t=10 0.0011456328
#> t=12 0.0013939562
plot(cph_predict_parts_survshap)
 cph_predict_parts_survlime <- predict_parts(
cph_exp,
new_observation = veteran[1, -c(3, 4)],
type = "survlime"
)
head(cph_predict_parts_survlime$result)
#> trt karno diagtime age prior celltypelarge
#> 1 0.003629477 -0.0007383551 8.119279e-05 0.0004152425 0.000347701 -0.007316818
#> celltypesmallcell celltypesquamous
#> 1 0.005774009 -0.005378658
plot(cph_predict_parts_survlime, type = "local_importance")
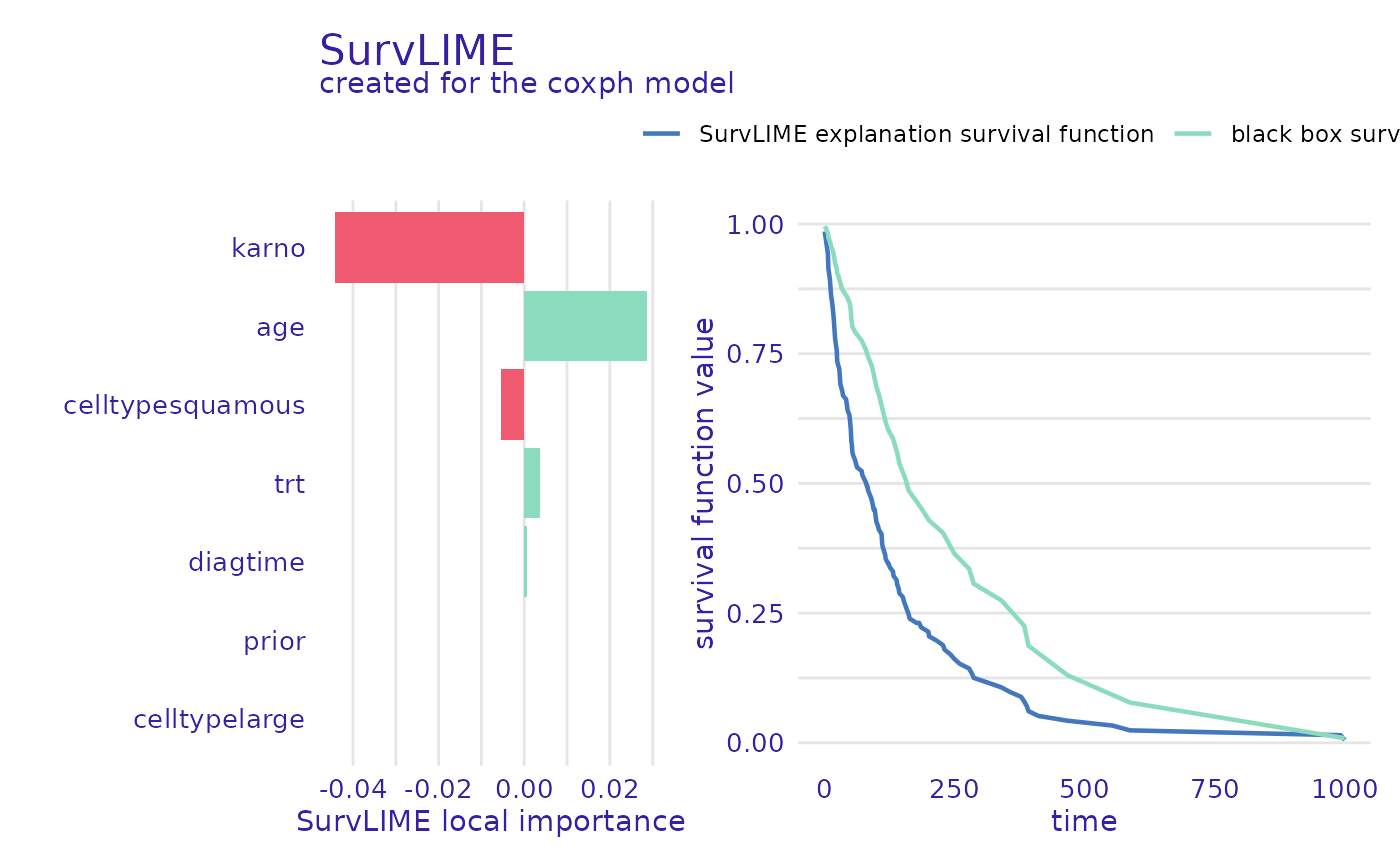
cph_predict_parts_survlime <- predict_parts(
cph_exp,
new_observation = veteran[1, -c(3, 4)],
type = "survlime"
)
head(cph_predict_parts_survlime$result)
#> trt karno diagtime age prior celltypelarge
#> 1 0.003629477 -0.0007383551 8.119279e-05 0.0004152425 0.000347701 -0.007316818
#> celltypesmallcell celltypesquamous
#> 1 0.005774009 -0.005378658
plot(cph_predict_parts_survlime, type = "local_importance")
 # }
# }