Instance Level Variable Attributions as Break Down, SHAP, aggregated SHAP or Oscillations explanations.
Model prediction is decomposed into parts that are attributed for particular variables.
From DALEX version 1.0 this function calls the break_down or
shap functions from the iBreakDown package or
ceteris_paribus from the ingredients package or
kernelshap from the kernelshap package.
Find information how to use the break_down method here: https://ema.drwhy.ai/breakDown.html.
Find information how to use the shap method here: https://ema.drwhy.ai/shapley.html.
Find information how to use the oscillations method here: https://ema.drwhy.ai/ceterisParibusOscillations.html.
Find information how to use the kernelshap method here: https://modeloriented.github.io/kernelshap/
aSHAP method provides explanations for a set of observations based on SHAP.
predict_parts(
explainer,
new_observation,
...,
N = if (substr(type, 1, 4) == "osci") 500 else NULL,
type = "break_down"
)
predict_parts_oscillations(explainer, new_observation, ...)
predict_parts_oscillations_uni(
explainer,
new_observation,
variable_splits_type = "uniform",
...
)
predict_parts_oscillations_emp(
explainer,
new_observation,
variable_splits = NULL,
variables = colnames(explainer$data),
...
)
predict_parts_break_down(explainer, new_observation, ...)
predict_parts_break_down_interactions(explainer, new_observation, ...)
predict_parts_shap(explainer, new_observation, ...)
predict_parts_shap_aggregated(explainer, new_observation, ...)
predict_parts_kernel_shap(explainer, new_observation, ...)
predict_parts_kernel_shap_break_down(explainer, new_observation, ...)
predict_parts_kernel_shap_aggreagted(explainer, new_observation, ...)
variable_attribution(
explainer,
new_observation,
...,
N = if (substr(type, 1, 4) == "osci") 500 else NULL,
type = "break_down"
)Arguments
- explainer
a model to be explained, preprocessed by the
explainfunction- new_observation
a new observation for which predictions need to be explained
- ...
other parameters that will be passed to
iBreakDown::break_down- N
the maximum number of observations used for calculation of attributions. By default NULL (use all) or 500 (for oscillations).
- type
the type of variable attributions. Either
shap,aggregated_shap,oscillations,oscillations_uni,oscillations_emp,break_down,break_down_interactions,kernel_shap,kernel_shap_break_downorkernel_shap_aggregated.- variable_splits_type
how variable grids shall be calculated? Will be passed to
ceteris_paribus.- variable_splits
named list of splits for variables. It is used by oscillations based measures. Will be passed to
ceteris_paribus.- variables
names of variables for which splits shall be calculated. Will be passed to
ceteris_paribus.
Value
Depending on the type there are different classes of the resulting object.
It's a data frame with calculated average response.
References
Explanatory Model Analysis. Explore, Explain, and Examine Predictive Models. https://ema.drwhy.ai/
Examples
library(DALEX)
new_dragon <- data.frame(
year_of_birth = 200,
height = 80,
weight = 12.5,
scars = 0,
number_of_lost_teeth = 5
)
model_lm <- lm(life_length ~ year_of_birth + height +
weight + scars + number_of_lost_teeth,
data = dragons)
explainer_lm <- explain(model_lm,
data = dragons,
y = dragons$year_of_birth,
label = "model_lm")
#> Preparation of a new explainer is initiated
#> -> model label : model_lm
#> -> data : 2000 rows 8 cols
#> -> target variable : 2000 values
#> -> predict function : yhat.lm will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package stats , ver. 4.2.3 , task regression ( default )
#> -> predicted values : numerical, min = 541.1056 , mean = 1370.986 , max = 3928.189
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -5421.316 , mean = -1450.523 , max = 1176.912
#> A new explainer has been created!
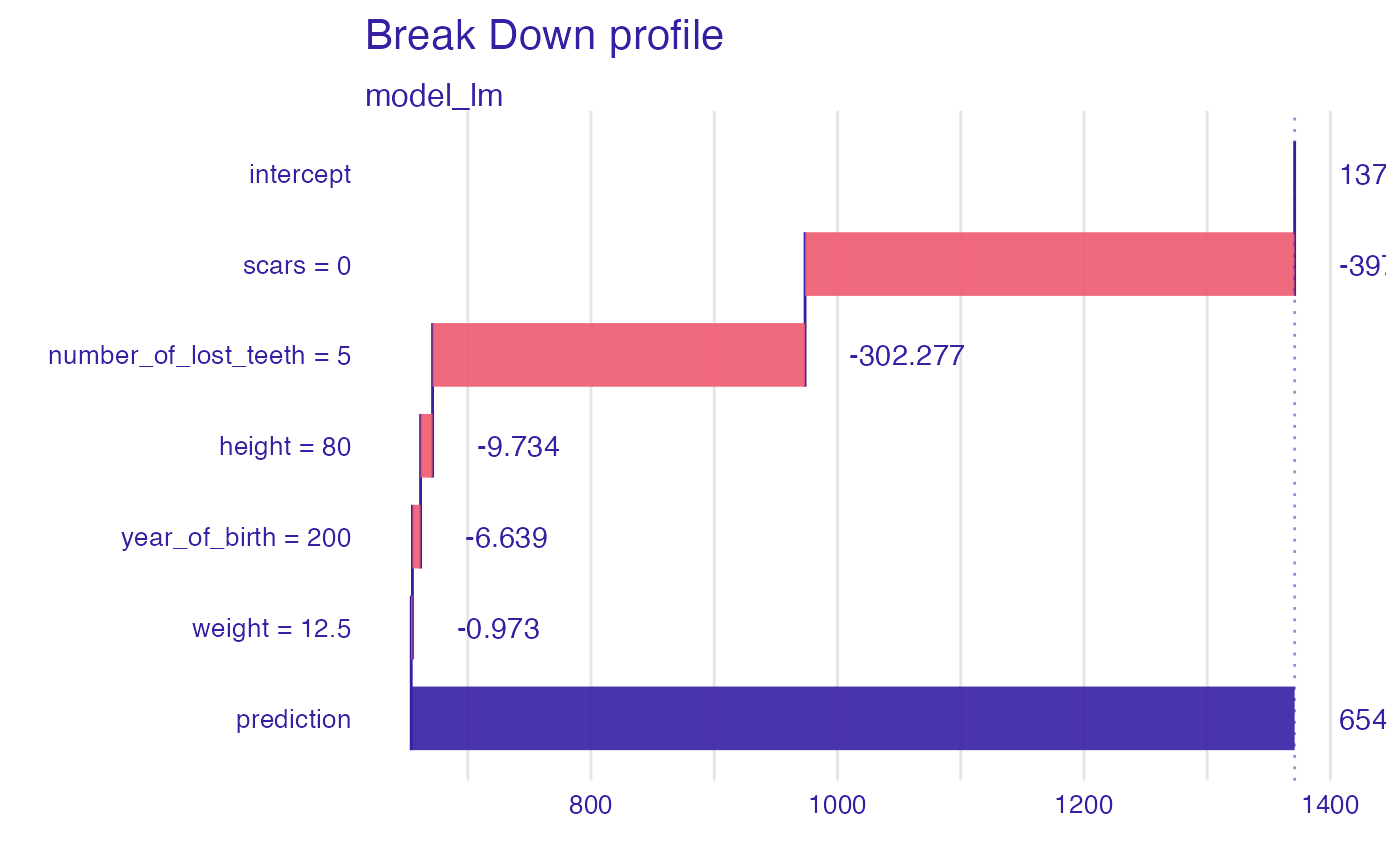
bd_lm <- predict_parts_break_down(explainer_lm, new_observation = new_dragon)
head(bd_lm)
#> contribution
#> model_lm: intercept 1370.986
#> model_lm: scars = 0 -397.300
#> model_lm: number_of_lost_teeth = 5 -302.277
#> model_lm: height = 80 -9.734
#> model_lm: year_of_birth = 200 -6.639
#> model_lm: weight = 12.5 -0.973
plot(bd_lm)
 # \donttest{
library("ranger")
model_ranger <- ranger(life_length ~ year_of_birth + height +
weight + scars + number_of_lost_teeth,
data = dragons, num.trees = 50)
explainer_ranger <- explain(model_ranger,
data = dragons,
y = dragons$year_of_birth,
label = "model_ranger")
#> Preparation of a new explainer is initiated
#> -> model label : model_ranger
#> -> data : 2000 rows 8 cols
#> -> target variable : 2000 values
#> -> predict function : yhat.ranger will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package ranger , ver. 0.14.1 , task regression ( default )
#> -> predicted values : numerical, min = 585.2637 , mean = 1370.777 , max = 3390.243
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -4961.243 , mean = -1450.314 , max = 1052.711
#> A new explainer has been created!
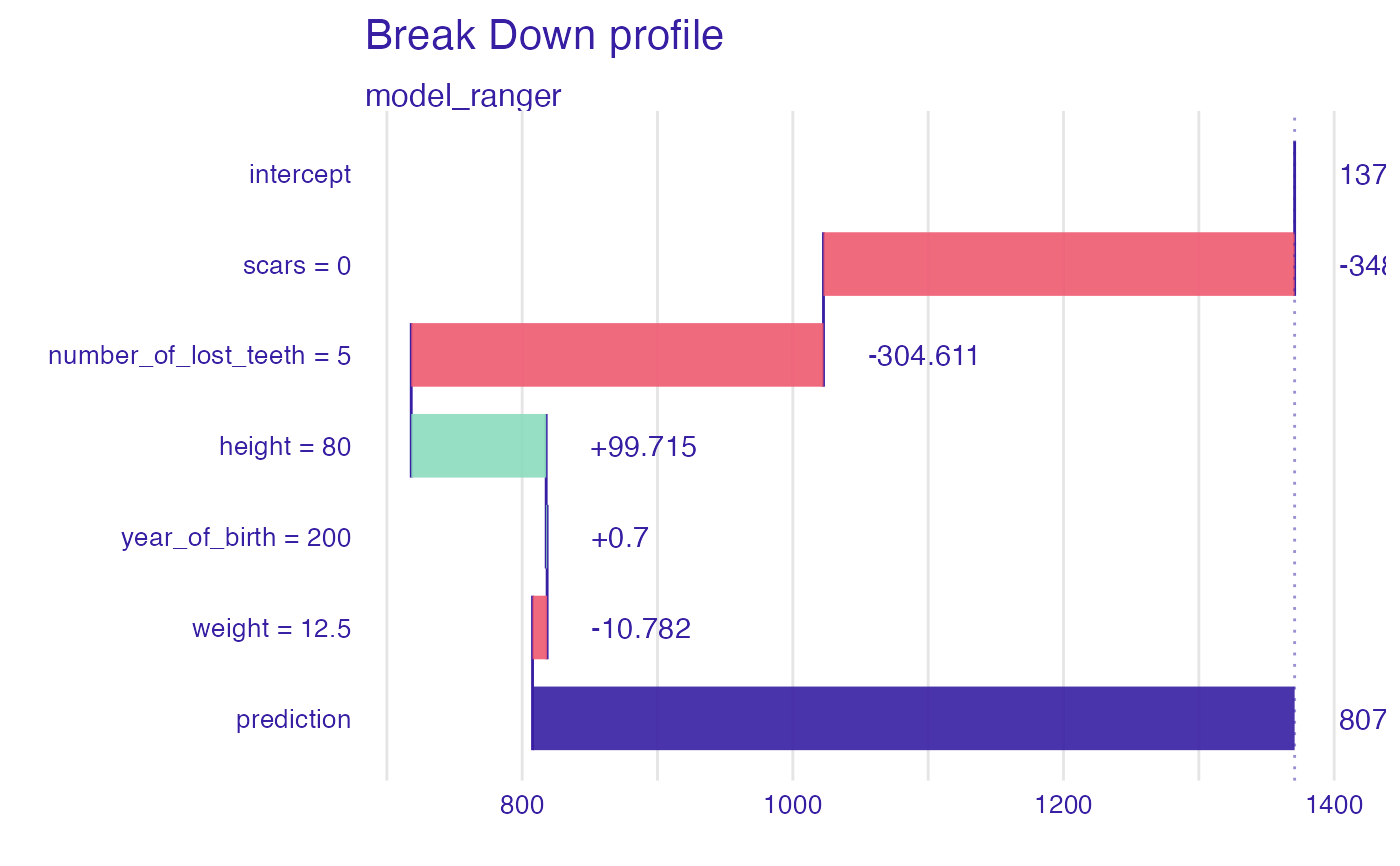
bd_ranger <- predict_parts_break_down(explainer_ranger, new_observation = new_dragon)
head(bd_ranger)
#> contribution
#> model_ranger: intercept 1370.777
#> model_ranger: scars = 0 -348.130
#> model_ranger: number_of_lost_teeth = 5 -304.611
#> model_ranger: height = 80 99.715
#> model_ranger: year_of_birth = 200 0.700
#> model_ranger: weight = 12.5 -10.782
plot(bd_ranger)
# \donttest{
library("ranger")
model_ranger <- ranger(life_length ~ year_of_birth + height +
weight + scars + number_of_lost_teeth,
data = dragons, num.trees = 50)
explainer_ranger <- explain(model_ranger,
data = dragons,
y = dragons$year_of_birth,
label = "model_ranger")
#> Preparation of a new explainer is initiated
#> -> model label : model_ranger
#> -> data : 2000 rows 8 cols
#> -> target variable : 2000 values
#> -> predict function : yhat.ranger will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package ranger , ver. 0.14.1 , task regression ( default )
#> -> predicted values : numerical, min = 585.2637 , mean = 1370.777 , max = 3390.243
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -4961.243 , mean = -1450.314 , max = 1052.711
#> A new explainer has been created!
bd_ranger <- predict_parts_break_down(explainer_ranger, new_observation = new_dragon)
head(bd_ranger)
#> contribution
#> model_ranger: intercept 1370.777
#> model_ranger: scars = 0 -348.130
#> model_ranger: number_of_lost_teeth = 5 -304.611
#> model_ranger: height = 80 99.715
#> model_ranger: year_of_birth = 200 0.700
#> model_ranger: weight = 12.5 -10.782
plot(bd_ranger)
 # }
# }