Dataset Level 2-Dimensional Variable Profile for Survival Models
Source:R/model_profile_2d.R
model_profile_2d.surv_explainer.RdThis function calculates explanations on a dataset level that help explore model response as a function of selected pairs of variables. The explanations are calculated as an extension of Partial Dependence Profiles or Accumulated Local Effects with the inclusion of the time dimension.
model_profile_2d(
explainer,
variables = NULL,
N = 100,
categorical_variables = NULL,
grid_points = 25,
center = FALSE,
variable_splits_type = "uniform",
type = "partial",
output_type = "survival"
)
# S3 method for surv_explainer
model_profile_2d(
explainer,
variables = NULL,
N = 100,
categorical_variables = NULL,
grid_points = 25,
center = FALSE,
variable_splits_type = "uniform",
type = "partial",
output_type = "survival"
)Arguments
- explainer
an explainer object - model preprocessed by the
explain()function- variables
list of character vectors of length 2, names of pairs of variables to be explained
- N
number of observations used for the calculation of aggregated profiles. By default
100. IfNULLall observations are used.- categorical_variables
character, a vector of names of additional variables which should be treated as categorical (factors are automatically treated as categorical variables). If it contains variable names not present in the
variablesargument, they will be added at the end.- grid_points
maximum number of points for profile calculations. Note that the final number of points may be lower than grid_points. Will be passed to internal function. By default
25.- center
logical, should profiles be centered around the average prediction
- variable_splits_type
character, decides how variable grids should be calculated. Use
"quantiles"for quantiles or"uniform"(default) to get uniform grid of points. Used only iftype = "partial".- type
the type of variable profile,
"partial"for Partial Dependence or"accumulated"for Accumulated Local Effects- output_type
either
"survival","chf"or"risk"the type of survival model output that should be considered for explanations. If"survival"the explanations are based on the survival function. If"chf"the explanations are based on the cumulative hazard function. Otherwise the scalar risk predictions are used by theDALEX::predict_profilefunction.
Value
An object of class model_profile_2d_survival. It is a list with the element result containing the results of the calculation.
Examples
# \donttest{
library(survival)
library(survex)
cph <- coxph(Surv(time, status) ~ ., data = veteran, model = TRUE, x = TRUE, y = TRUE)
cph_exp <- explain(cph)
#> Preparation of a new explainer is initiated
#> -> model label : coxph ( default )
#> -> data : 137 rows 6 cols ( extracted from the model )
#> -> target variable : 137 values ( 128 events and 9 censored , censoring rate = 0.066 ) ( extracted from the model )
#> -> times : 50 unique time points , min = 1.5 , median survival time = 80 , max = 999
#> -> times : ( generated from y as uniformly distributed survival quantiles based on Kaplan-Meier estimator )
#> -> predict function : predict.coxph with type = 'risk' will be used ( default )
#> -> predict survival function : predictSurvProb.coxph will be used ( default )
#> -> predict cumulative hazard function : -log(predict_survival_function) will be used ( default )
#> -> model_info : package survival , ver. 3.7.0 , task survival ( default )
#> A new explainer has been created!
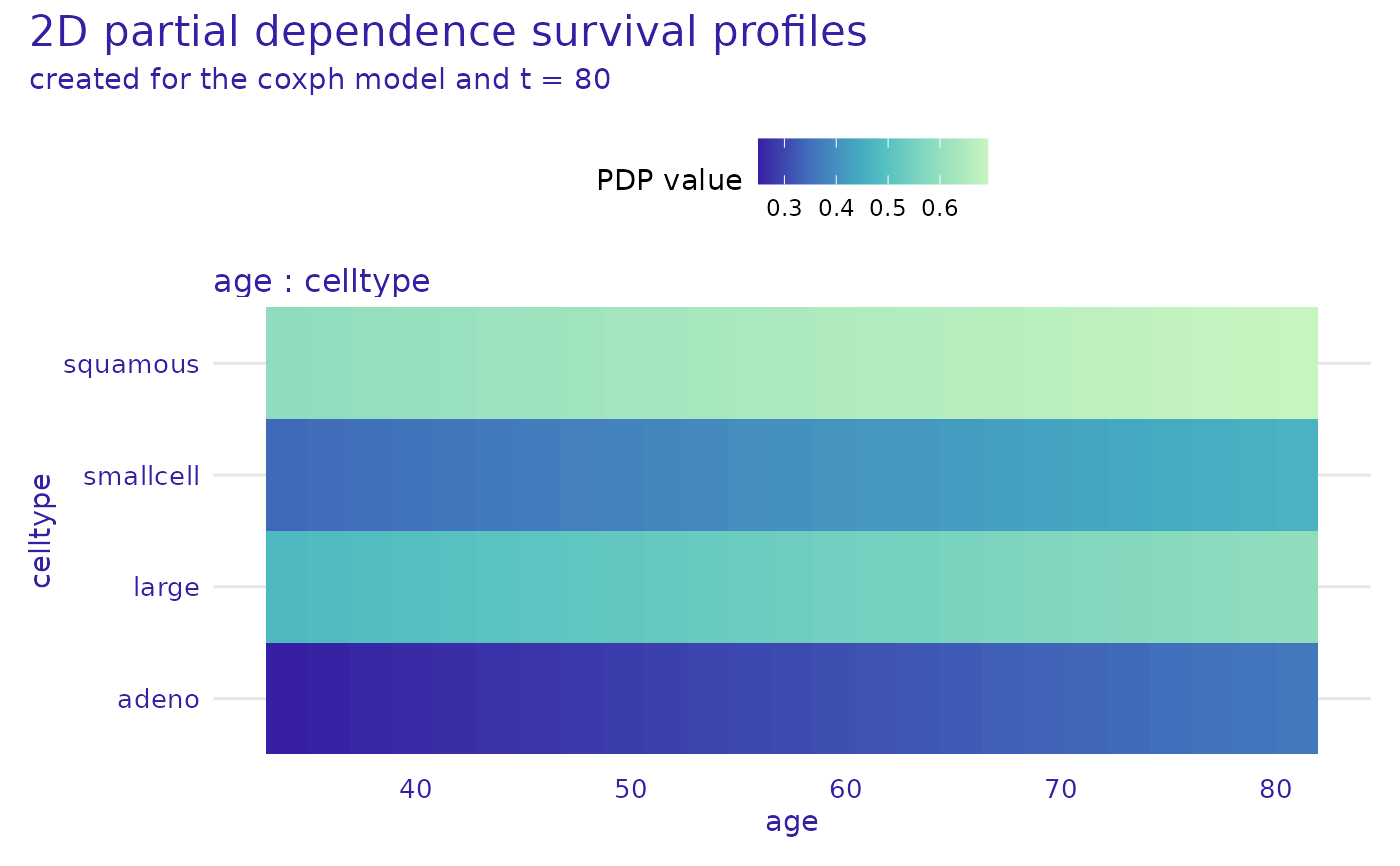
cph_model_profile_2d <- model_profile_2d(cph_exp,
variables = list(c("age", "celltype"))
)
head(cph_model_profile_2d$result)
#> _v1name_ _v2name_ _v1type_ _v2type_ _v1value_ _v2value_ _times_
#> 1 age celltype numerical categorical 34 adeno 1.5
#> 2 age celltype numerical categorical 35.9583333333333 adeno 1.5
#> 3 age celltype numerical categorical 37.9166666666667 adeno 1.5
#> 4 age celltype numerical categorical 39.875 adeno 1.5
#> 5 age celltype numerical categorical 41.8333333333333 adeno 1.5
#> 6 age celltype numerical categorical 43.7916666666667 adeno 1.5
#> _label_ _yhat_
#> 1 coxph 0.9707866
#> 2 coxph 0.9712691
#> 3 coxph 0.9717438
#> 4 coxph 0.9722109
#> 5 coxph 0.9726704
#> 6 coxph 0.9731225
plot(cph_model_profile_2d)
#> Warning: Plot will be prepared for the median survial time. For another time point, set the value of `times`.
 cph_model_profile_2d_ale <- model_profile_2d(cph_exp,
variables = list(c("age", "karno")),
type = "accumulated"
)
head(cph_model_profile_2d_ale$result)
#> _v1name_ _v2name_ _v1type_ _v2type_ _v1value_ _v2value_ _times_ _yhat_
#> 1 age karno numerical numerical 35 20 1.5 0.9844186
#> 2 age karno numerical numerical 35 20 4.0 0.9614581
#> 3 age karno numerical numerical 35 20 7.0 0.9386648
#> 4 age karno numerical numerical 35 20 8.0 0.9086675
#> 5 age karno numerical numerical 35 20 10.0 0.8939623
#> 6 age karno numerical numerical 35 20 12.0 0.8725289
#> _right_ _left_ _top_ _bottom_ _count_ _label_
#> 1 36.5 35 20 25 0 coxph
#> 2 36.5 35 20 25 0 coxph
#> 3 36.5 35 20 25 0 coxph
#> 4 36.5 35 20 25 0 coxph
#> 5 36.5 35 20 25 0 coxph
#> 6 36.5 35 20 25 0 coxph
plot(cph_model_profile_2d_ale)
#> Warning: Plot will be prepared for the median survial time. For another time point, set the value of `times`.
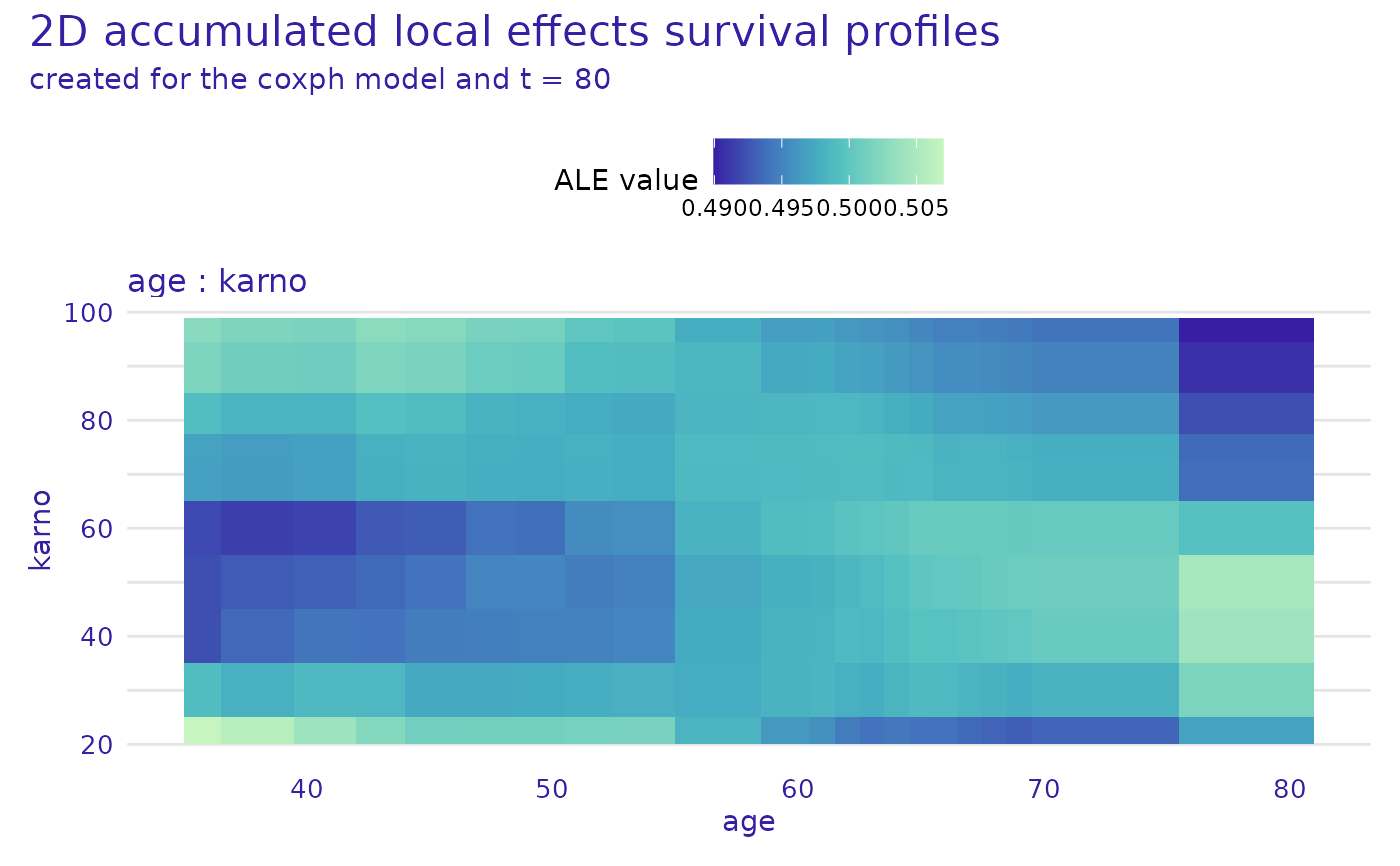
cph_model_profile_2d_ale <- model_profile_2d(cph_exp,
variables = list(c("age", "karno")),
type = "accumulated"
)
head(cph_model_profile_2d_ale$result)
#> _v1name_ _v2name_ _v1type_ _v2type_ _v1value_ _v2value_ _times_ _yhat_
#> 1 age karno numerical numerical 35 20 1.5 0.9844186
#> 2 age karno numerical numerical 35 20 4.0 0.9614581
#> 3 age karno numerical numerical 35 20 7.0 0.9386648
#> 4 age karno numerical numerical 35 20 8.0 0.9086675
#> 5 age karno numerical numerical 35 20 10.0 0.8939623
#> 6 age karno numerical numerical 35 20 12.0 0.8725289
#> _right_ _left_ _top_ _bottom_ _count_ _label_
#> 1 36.5 35 20 25 0 coxph
#> 2 36.5 35 20 25 0 coxph
#> 3 36.5 35 20 25 0 coxph
#> 4 36.5 35 20 25 0 coxph
#> 5 36.5 35 20 25 0 coxph
#> 6 36.5 35 20 25 0 coxph
plot(cph_model_profile_2d_ale)
#> Warning: Plot will be prepared for the median survial time. For another time point, set the value of `times`.
 # }
# }