This function calculated individual profiles aka Ceteris Paribus Profiles.
From DALEX version 1.0 this function calls the ceteris_paribus from the ingredients package.
Find information how to use this function here: https://ema.drwhy.ai/ceterisParibus.html.
predict_profile(
explainer,
new_observation,
variables = NULL,
...,
type = "ceteris_paribus",
variable_splits_type = "uniform"
)
individual_profile(
explainer,
new_observation,
variables = NULL,
...,
type = "ceteris_paribus",
variable_splits_type = "uniform"
)Arguments
- explainer
a model to be explained, preprocessed by the
explainfunction- new_observation
a new observation for which predictions need to be explained
- variables
character - names of variables to be explained
- ...
other parameters
- type
character, currently only the
ceteris_paribusis implemented- variable_splits_type
how variable grids shall be calculated? Use "quantiles" (default) for percentiles or "uniform" to get uniform grid of points. Will be passed to `ingredients`.
Value
An object of the class ceteris_paribus_explainer.
It's a data frame with calculated average response.
References
Explanatory Model Analysis. Explore, Explain, and Examine Predictive Models. https://ema.drwhy.ai/
Examples
new_dragon <- data.frame(year_of_birth = 200,
height = 80,
weight = 12.5,
scars = 0,
number_of_lost_teeth = 5)
dragon_lm_model4 <- lm(life_length ~ year_of_birth + height +
weight + scars + number_of_lost_teeth,
data = dragons)
dragon_lm_explainer4 <- explain(dragon_lm_model4, data = dragons, y = dragons$year_of_birth,
label = "model_4v")
#> Preparation of a new explainer is initiated
#> -> model label : model_4v
#> -> data : 2000 rows 8 cols
#> -> target variable : 2000 values
#> -> predict function : yhat.lm will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package stats , ver. 4.2.3 , task regression ( default )
#> -> predicted values : numerical, min = 541.1056 , mean = 1370.986 , max = 3928.189
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -5421.316 , mean = -1450.523 , max = 1176.912
#> A new explainer has been created!
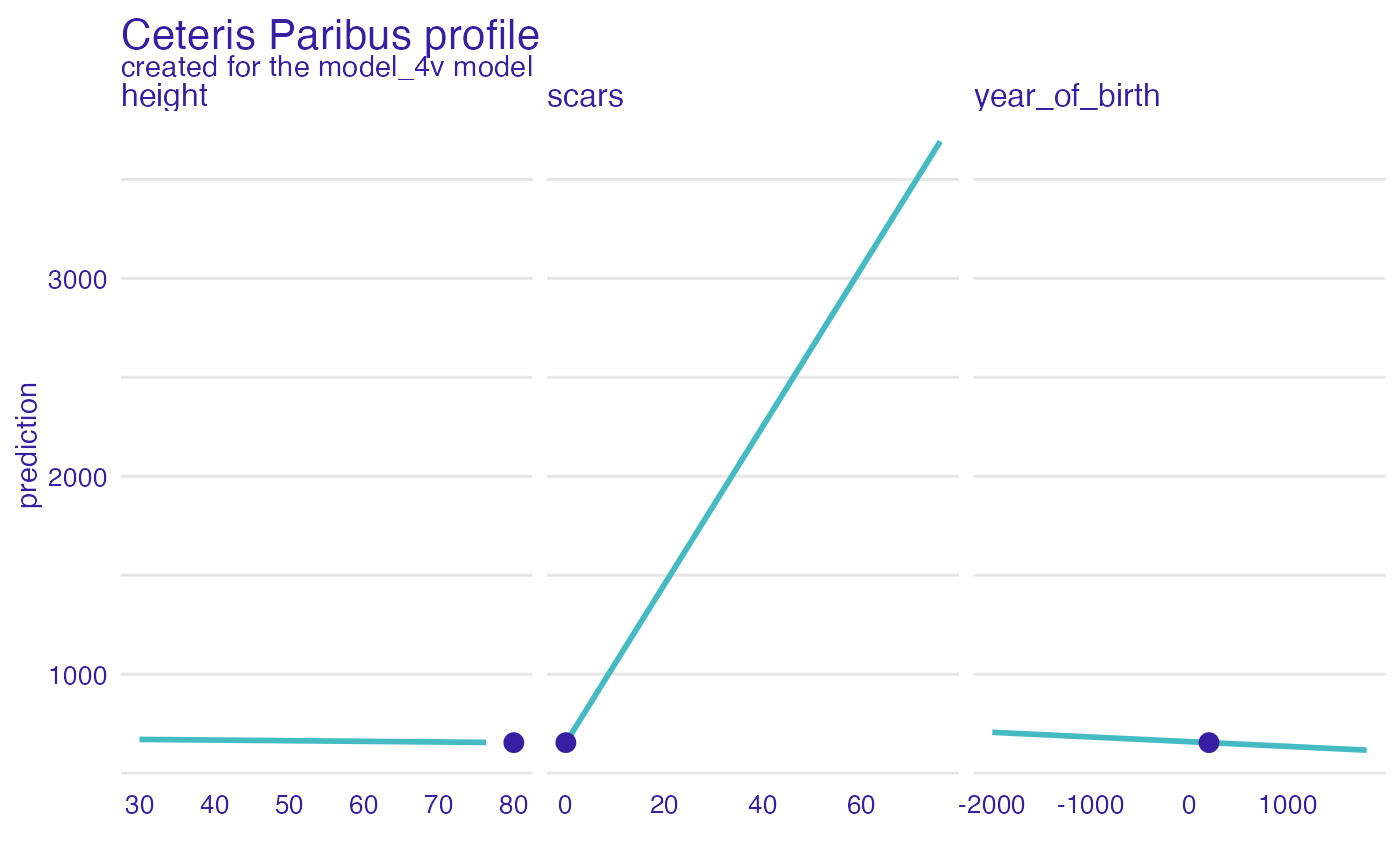
dragon_lm_predict4 <- predict_profile(dragon_lm_explainer4,
new_observation = new_dragon,
variables = c("year_of_birth", "height", "scars"))
head(dragon_lm_predict4)
#> Top profiles :
#> year_of_birth height weight scars number_of_lost_teeth _yhat_
#> 1 -1999.00 80 12.5 0 5 706.2869
#> 1.1 -1961.01 80 12.5 0 5 705.3847
#> 1.2 -1923.02 80 12.5 0 5 704.4825
#> 1.3 -1885.03 80 12.5 0 5 703.5803
#> 1.4 -1847.04 80 12.5 0 5 702.6780
#> 1.5 -1809.05 80 12.5 0 5 701.7758
#> _vname_ _ids_ _label_
#> 1 year_of_birth 1 model_4v
#> 1.1 year_of_birth 1 model_4v
#> 1.2 year_of_birth 1 model_4v
#> 1.3 year_of_birth 1 model_4v
#> 1.4 year_of_birth 1 model_4v
#> 1.5 year_of_birth 1 model_4v
#>
#>
#> Top observations:
#> year_of_birth height weight scars number_of_lost_teeth _yhat_ _label_
#> 1 200 80 12.5 0 5 654.0636 model_4v
#> _ids_
#> 1 1
plot(dragon_lm_predict4,
variables = c("year_of_birth", "height", "scars"))
 # \donttest{
library("ranger")
dragon_ranger_model4 <- ranger(life_length ~ year_of_birth + height +
weight + scars + number_of_lost_teeth,
data = dragons, num.trees = 50)
dragon_ranger_explainer4 <- explain(dragon_ranger_model4, data = dragons, y = dragons$year_of_birth,
label = "model_ranger")
#> Preparation of a new explainer is initiated
#> -> model label : model_ranger
#> -> data : 2000 rows 8 cols
#> -> target variable : 2000 values
#> -> predict function : yhat.ranger will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package ranger , ver. 0.14.1 , task regression ( default )
#> -> predicted values : numerical, min = 587.5616 , mean = 1370.765 , max = 3566.677
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -4986.952 , mean = -1450.302 , max = 1029.847
#> A new explainer has been created!
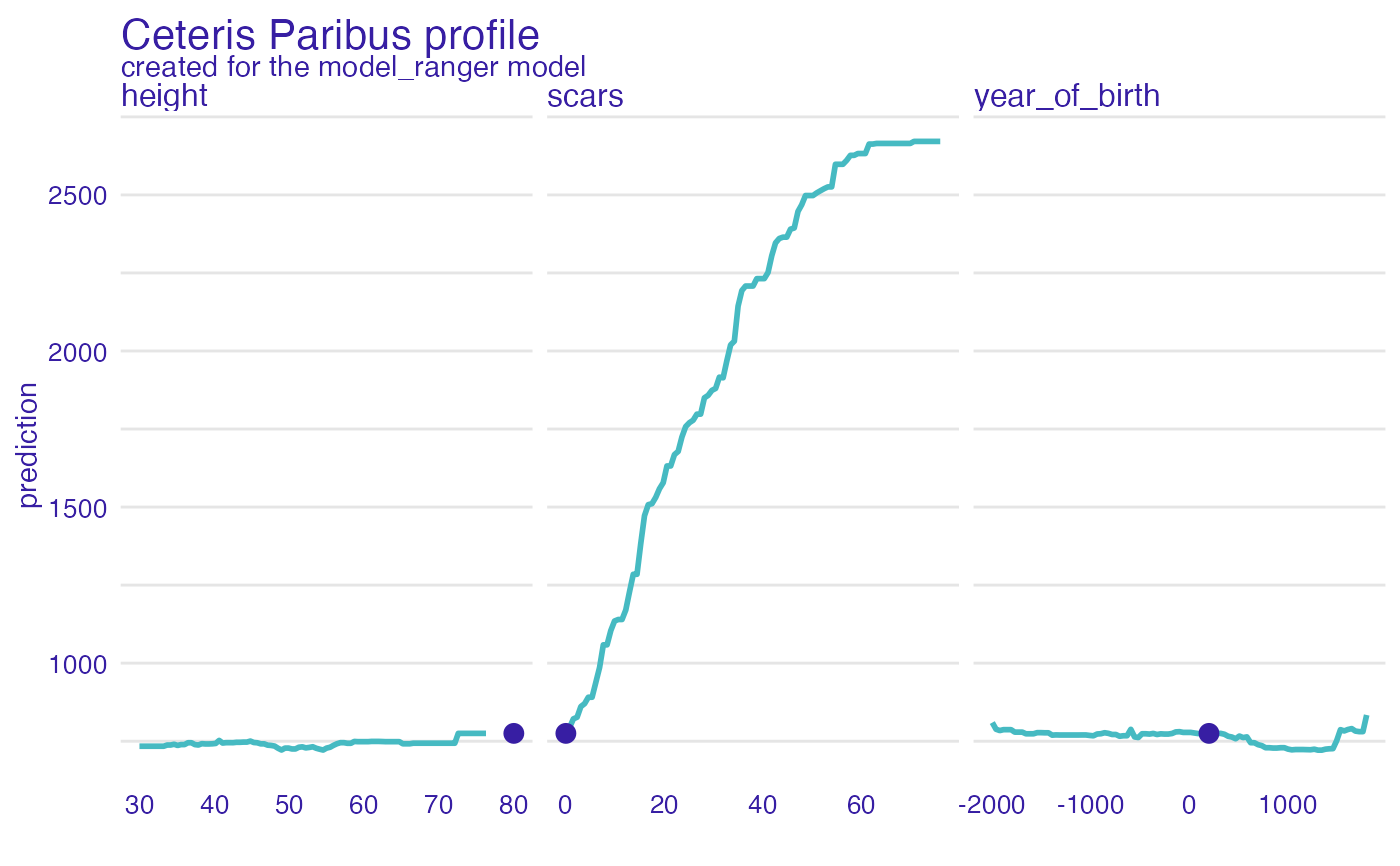
dragon_ranger_predict4 <- predict_profile(dragon_ranger_explainer4,
new_observation = new_dragon,
variables = c("year_of_birth", "height", "scars"))
head(dragon_ranger_predict4)
#> Top profiles :
#> year_of_birth height weight scars number_of_lost_teeth _yhat_
#> 1 -1999.00 80 12.5 0 5 809.6815
#> 1.1 -1961.01 80 12.5 0 5 788.3777
#> 1.2 -1923.02 80 12.5 0 5 784.1321
#> 1.3 -1885.03 80 12.5 0 5 786.9855
#> 1.4 -1847.04 80 12.5 0 5 786.9855
#> 1.5 -1809.05 80 12.5 0 5 786.9855
#> _vname_ _ids_ _label_
#> 1 year_of_birth 1 model_ranger
#> 1.1 year_of_birth 1 model_ranger
#> 1.2 year_of_birth 1 model_ranger
#> 1.3 year_of_birth 1 model_ranger
#> 1.4 year_of_birth 1 model_ranger
#> 1.5 year_of_birth 1 model_ranger
#>
#>
#> Top observations:
#> year_of_birth height weight scars number_of_lost_teeth _yhat_ _label_
#> 1 200 80 12.5 0 5 775.0935 model_ranger
#> _ids_
#> 1 1
plot(dragon_ranger_predict4,
variables = c("year_of_birth", "height", "scars"))
# \donttest{
library("ranger")
dragon_ranger_model4 <- ranger(life_length ~ year_of_birth + height +
weight + scars + number_of_lost_teeth,
data = dragons, num.trees = 50)
dragon_ranger_explainer4 <- explain(dragon_ranger_model4, data = dragons, y = dragons$year_of_birth,
label = "model_ranger")
#> Preparation of a new explainer is initiated
#> -> model label : model_ranger
#> -> data : 2000 rows 8 cols
#> -> target variable : 2000 values
#> -> predict function : yhat.ranger will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package ranger , ver. 0.14.1 , task regression ( default )
#> -> predicted values : numerical, min = 587.5616 , mean = 1370.765 , max = 3566.677
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -4986.952 , mean = -1450.302 , max = 1029.847
#> A new explainer has been created!
dragon_ranger_predict4 <- predict_profile(dragon_ranger_explainer4,
new_observation = new_dragon,
variables = c("year_of_birth", "height", "scars"))
head(dragon_ranger_predict4)
#> Top profiles :
#> year_of_birth height weight scars number_of_lost_teeth _yhat_
#> 1 -1999.00 80 12.5 0 5 809.6815
#> 1.1 -1961.01 80 12.5 0 5 788.3777
#> 1.2 -1923.02 80 12.5 0 5 784.1321
#> 1.3 -1885.03 80 12.5 0 5 786.9855
#> 1.4 -1847.04 80 12.5 0 5 786.9855
#> 1.5 -1809.05 80 12.5 0 5 786.9855
#> _vname_ _ids_ _label_
#> 1 year_of_birth 1 model_ranger
#> 1.1 year_of_birth 1 model_ranger
#> 1.2 year_of_birth 1 model_ranger
#> 1.3 year_of_birth 1 model_ranger
#> 1.4 year_of_birth 1 model_ranger
#> 1.5 year_of_birth 1 model_ranger
#>
#>
#> Top observations:
#> year_of_birth height weight scars number_of_lost_teeth _yhat_ _label_
#> 1 200 80 12.5 0 5 775.0935 model_ranger
#> _ids_
#> 1 1
plot(dragon_ranger_predict4,
variables = c("year_of_birth", "height", "scars"))
 # }
# }