Plot Instance Level Residual Diagnostics
Source:R/plot_predict_diagnostics.R
plot.predict_diagnostics.RdPlot Instance Level Residual Diagnostics
# S3 method for predict_diagnostics
plot(x, ...)Arguments
- x
an object with instance level residual diagnostics created with
predict_diagnosticsfunction- ...
other parameters that will be passed to
plot.ceteris_paribus_explaine.
Value
an ggplot2 object of the class gg.
Examples
# \donttest{
library("ranger")
titanic_glm_model <- ranger(survived ~ gender + age + class + fare + sibsp + parch,
data = titanic_imputed)
explainer_glm <- explain(titanic_glm_model,
data = titanic_imputed,
y = titanic_imputed$survived)
#> Preparation of a new explainer is initiated
#> -> model label : ranger ( default )
#> -> data : 2207 rows 8 cols
#> -> target variable : 2207 values
#> -> predict function : yhat.ranger will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package ranger , ver. 0.14.1 , task regression ( default )
#> -> predicted values : numerical, min = 0.009358695 , mean = 0.3226251 , max = 0.9954396
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.8065088 , mean = -0.0004683031 , max = 0.8752794
#> A new explainer has been created!
johny_d <- titanic_imputed[24, c("gender", "age", "class", "fare", "sibsp", "parch")]
pl <- predict_diagnostics(explainer_glm, johny_d, variables = NULL)
#> Warning: p-value will be approximate in the presence of ties
plot(pl)
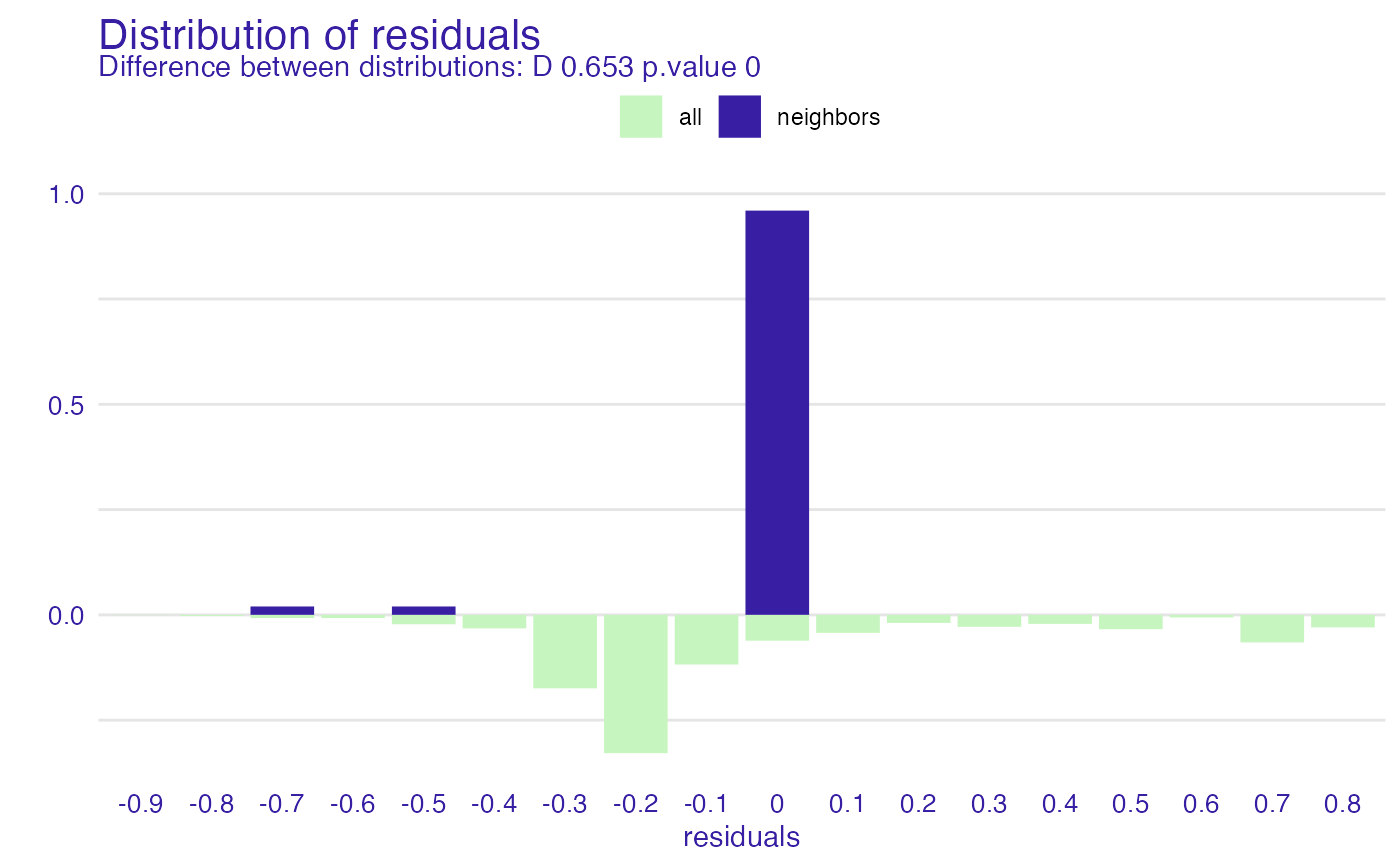 pl <- predict_diagnostics(explainer_glm, johny_d,
neighbors = 10,
variables = c("age", "fare"))
plot(pl)
pl <- predict_diagnostics(explainer_glm, johny_d,
neighbors = 10,
variables = c("age", "fare"))
plot(pl)
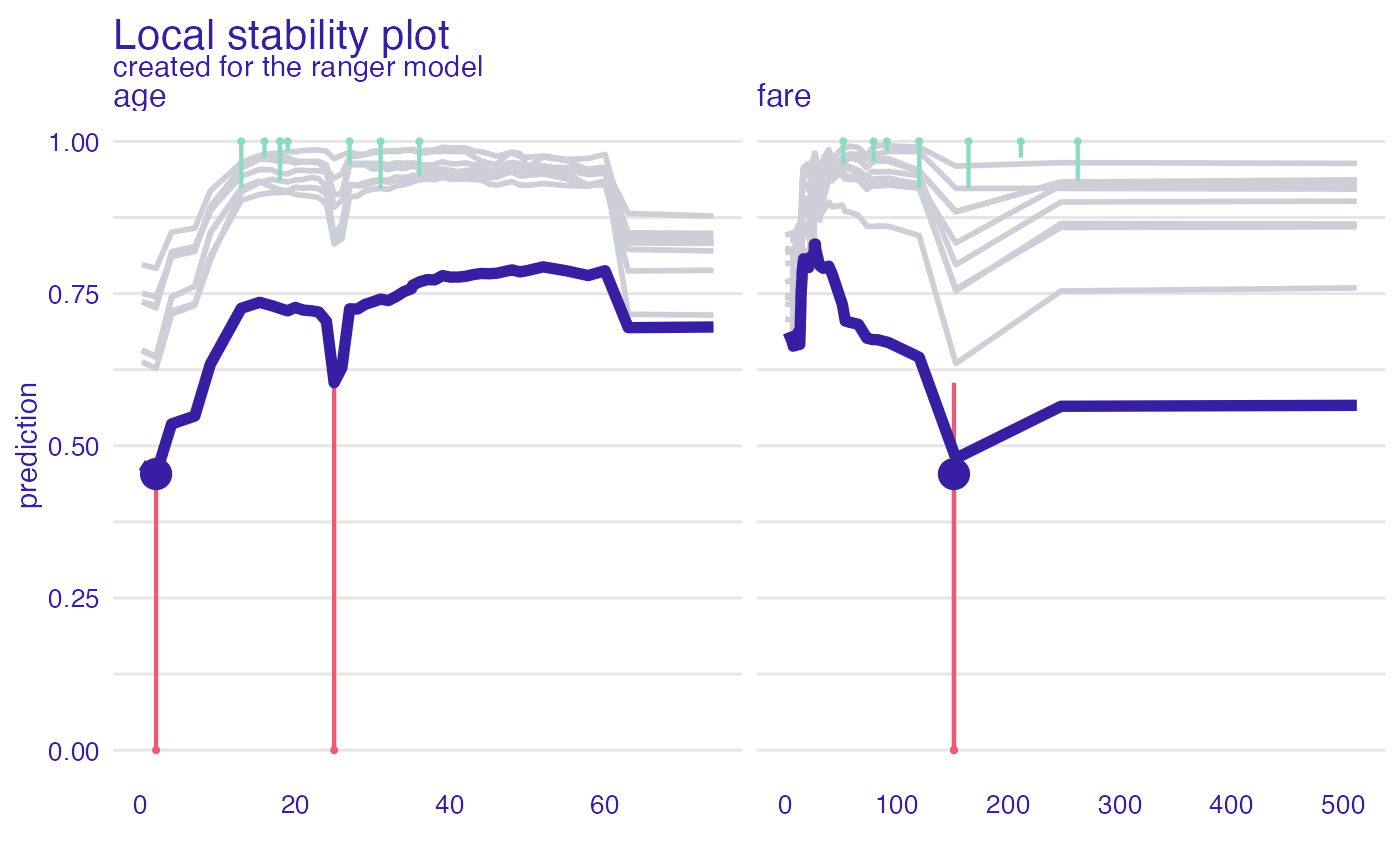 pl <- predict_diagnostics(explainer_glm,
johny_d,
neighbors = 10,
variables = c("class", "gender"))
plot(pl)
#> 'variable_type' changed to 'categorical' due to lack of numerical variables.
#> 'variable_type' changed to 'categorical' due to lack of numerical variables.
pl <- predict_diagnostics(explainer_glm,
johny_d,
neighbors = 10,
variables = c("class", "gender"))
plot(pl)
#> 'variable_type' changed to 'categorical' due to lack of numerical variables.
#> 'variable_type' changed to 'categorical' due to lack of numerical variables.
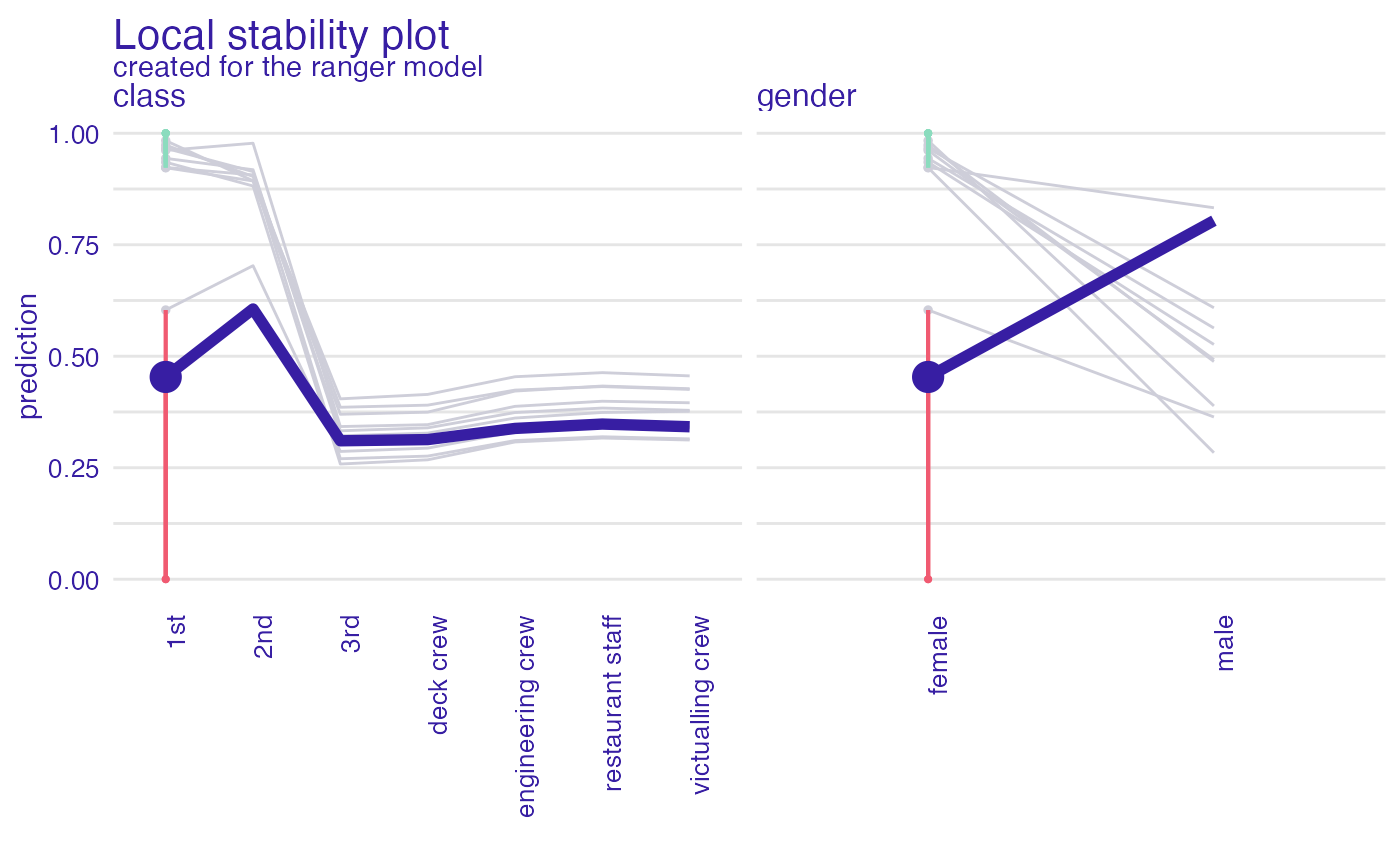 # }
# }