Plot List of Explanations
# S3 method for list
plot(x, ...)Arguments
- x
a list of explanations of the same class
- ...
other parameters
Value
An object of the class ggplot.
Examples
# \donttest{
library("ranger")
titanic_ranger_model <- ranger(survived~., data = titanic_imputed, num.trees = 50,
probability = TRUE)
explainer_ranger <- explain(titanic_ranger_model, data = titanic_imputed[,-8],
y = titanic_imputed$survived)
#> Preparation of a new explainer is initiated
#> -> model label : ranger ( default )
#> -> data : 2207 rows 7 cols
#> -> target variable : 2207 values
#> -> predict function : yhat.ranger will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package ranger , ver. 0.14.1 , task classification ( default )
#> -> predicted values : numerical, min = 0.0005555556 , mean = 0.3205157 , max = 0.9883577
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.831336 , mean = 0.001641051 , max = 0.8773243
#> A new explainer has been created!
mp_ranger <- model_performance(explainer_ranger)
titanic_ranger_model2 <- ranger(survived~gender + fare, data = titanic_imputed,
num.trees = 50, probability = TRUE)
explainer_ranger2 <- explain(titanic_ranger_model2, data = titanic_imputed[,-8],
y = titanic_imputed$survived,
label = "ranger2")
#> Preparation of a new explainer is initiated
#> -> model label : ranger2
#> -> data : 2207 rows 7 cols
#> -> target variable : 2207 values
#> -> predict function : yhat.ranger will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package ranger , ver. 0.14.1 , task classification ( default )
#> -> predicted values : numerical, min = 0.1572722 , mean = 0.3224717 , max = 0.9032262
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.8677249 , mean = -0.0003149388 , max = 0.8427278
#> A new explainer has been created!
mp_ranger2 <- model_performance(explainer_ranger2)
plot(list(mp_ranger, mp_ranger2), geom = "prc")
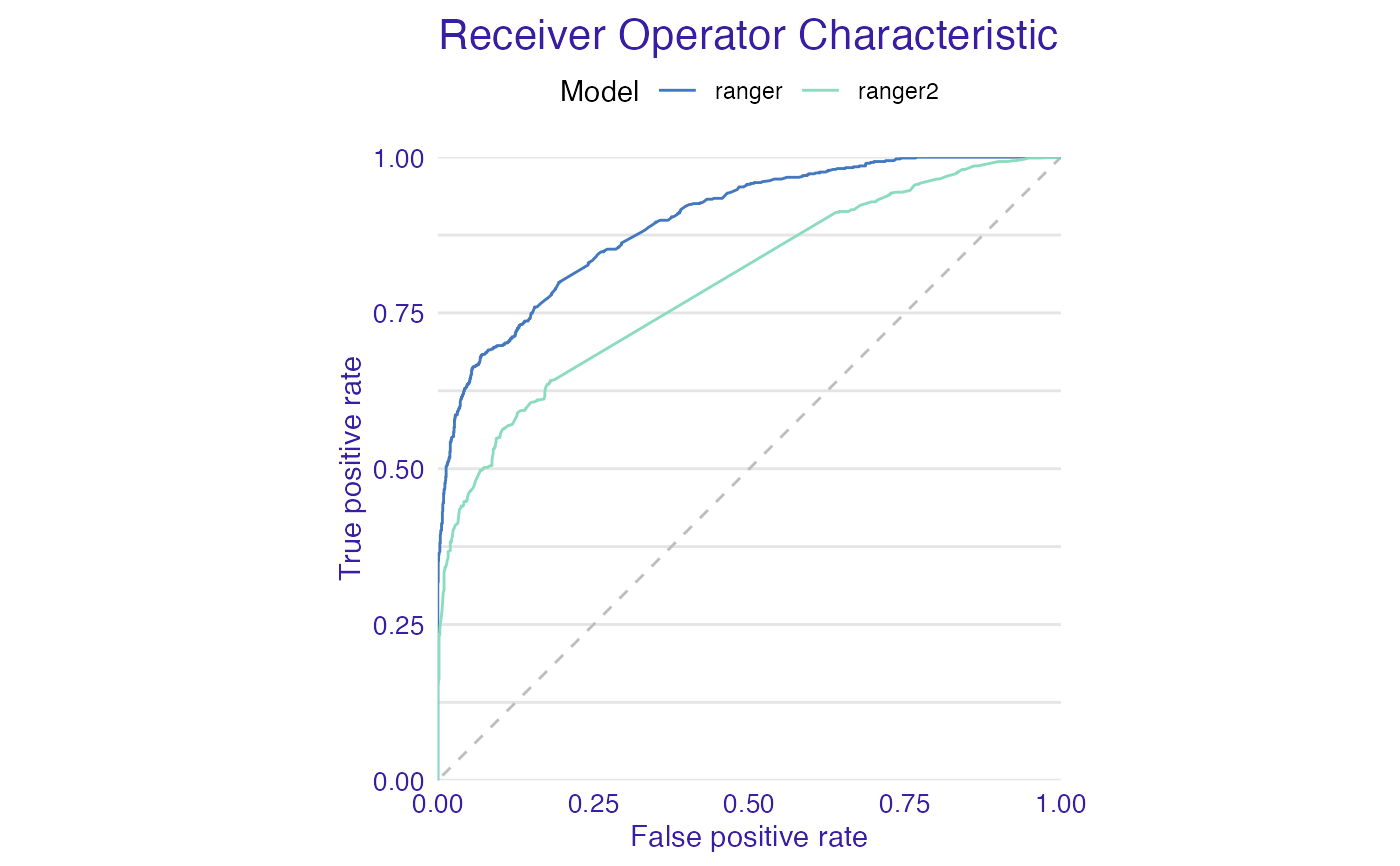
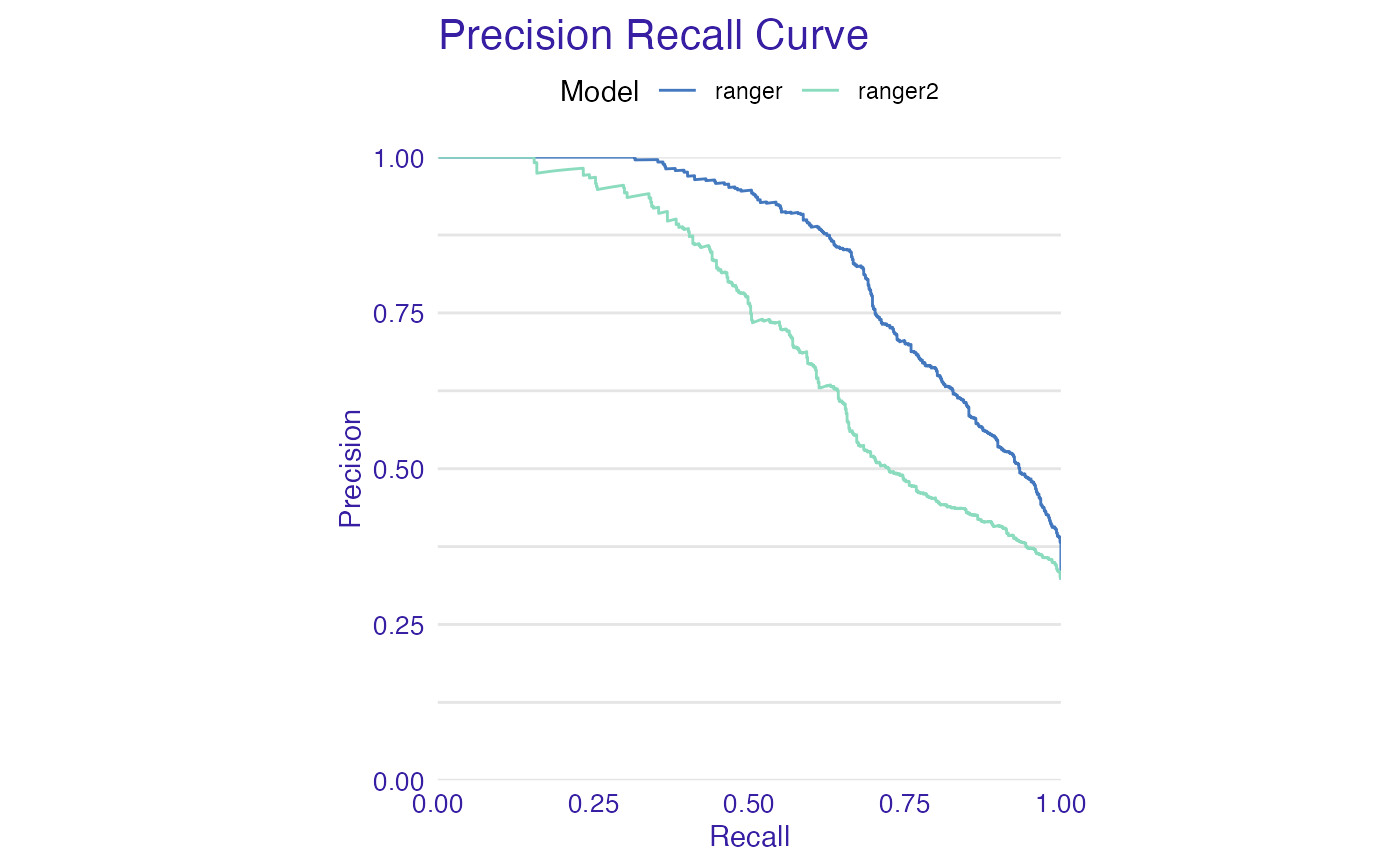 plot(list(mp_ranger, mp_ranger2), geom = "roc")
plot(list(mp_ranger, mp_ranger2), geom = "roc")
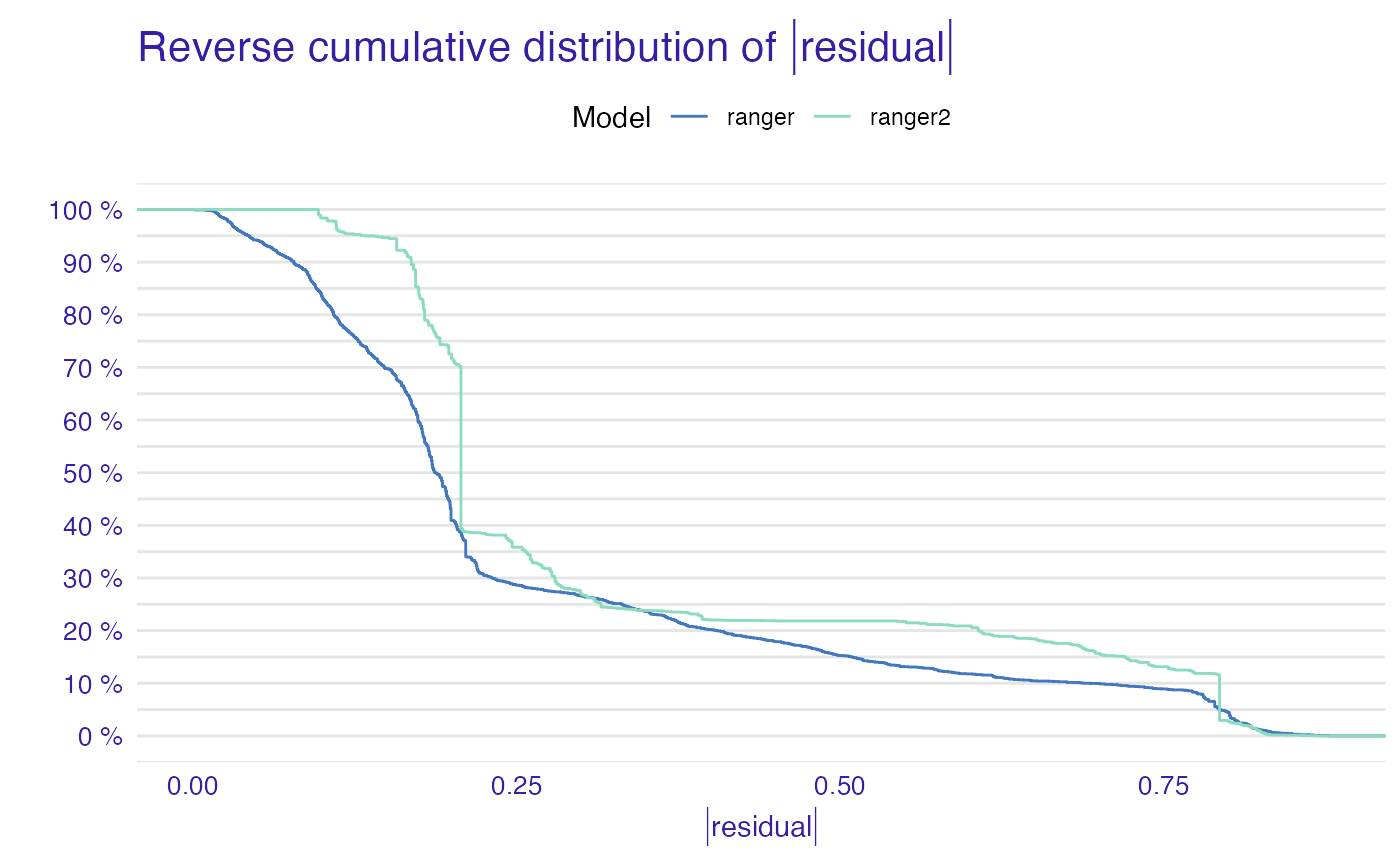
 tmp <- list(mp_ranger, mp_ranger2)
names(tmp) <- c("ranger", "ranger2")
plot(tmp)
tmp <- list(mp_ranger, mp_ranger2)
names(tmp) <- c("ranger", "ranger2")
plot(tmp)
 # }
# }