General introduction: iBreakDown plots for Sinking of the RMS Titanic
Przemyslaw Biecek
2022-02-06
Source:vignettes/vignette_iBreakDown_titanic.Rmd
vignette_iBreakDown_titanic.RmdData for Titanic survival
Let’s see an example for iBreakDown plots for survival probability of Titanic passengers. First, let’s see the data, we will find quite nice data from in the DALEX package (orginally stablelearner).
#> gender age class embarked country fare sibsp parch survived
#> 1 male 42 3rd Southampton United States 7.11 0 0 no
#> 2 male 13 3rd Southampton United States 20.05 0 2 no
#> 3 male 16 3rd Southampton United States 20.05 1 1 no
#> 4 female 39 3rd Southampton England 20.05 1 1 yes
#> 5 female 16 3rd Southampton Norway 7.13 0 0 yes
#> 6 male 25 3rd Southampton United States 7.13 0 0 yesModel for Titanic survival
Ok, now it’s time to create a model. Let’s use the Random Forest model.
# prepare model
library("randomForest")
titanic <- na.omit(titanic)
model_titanic_rf <- randomForest(survived == "yes" ~ gender + age + class + embarked +
fare + sibsp + parch, data = titanic)
model_titanic_rf#>
#> Call:
#> randomForest(formula = survived == "yes" ~ gender + age + class + embarked + fare + sibsp + parch, data = titanic)
#> Type of random forest: regression
#> Number of trees: 500
#> No. of variables tried at each split: 2
#>
#> Mean of squared residuals: 0.1425301
#> % Var explained: 34.97Explainer for Titanic survival
The third step (it’s optional but useful) is to create a DALEX explainer for Random Forest model.
library("DALEX")
explain_titanic_rf <- explain(model_titanic_rf,
data = titanic[,-9],
y = titanic$survived == "yes",
label = "Random Forest v7")#> Preparation of a new explainer is initiated
#> -> model label : Random Forest v7
#> -> data : 2099 rows 8 cols
#> -> target variable : 2099 values
#> -> predict function : yhat.randomForest will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package randomForest , ver. 4.7.1 , task regression ( default )
#> -> model_info : Model info detected regression task but 'y' is a logical . ( WARNING )
#> -> model_info : By deafult regressions tasks supports only numercical 'y' parameter.
#> -> model_info : Consider changing to numerical vector.
#> -> model_info : Otherwise I will not be able to calculate residuals or loss function.
#> -> predicted values : numerical, min = 0.01000576 , mean = 0.3249614 , max = 0.9917294
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.7829437 , mean = -0.000521225 , max = 0.8976734
#> A new explainer has been created! Break Down plot with D3
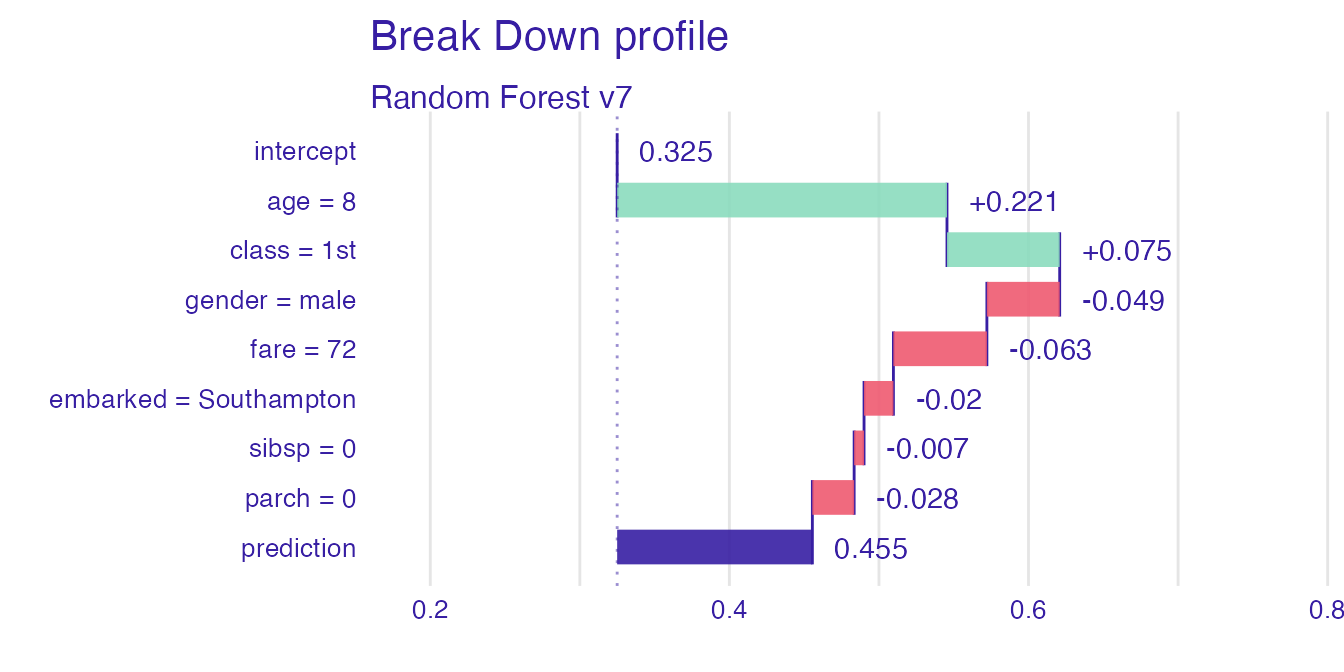
Let’s see Break Down for model predictions for 8 years old male from 1st class that embarked from port C.
new_passanger <- data.frame(
class = factor("1st", levels = c("1st", "2nd", "3rd", "deck crew", "engineering crew", "restaurant staff", "victualling crew")),
gender = factor("male", levels = c("female", "male")),
age = 8,
sibsp = 0,
parch = 0,
fare = 72,
embarked = factor("Southampton", levels = c("Belfast", "Cherbourg", "Queenstown", "Southampton"))
)Calculate variable attributions
library("iBreakDown")
rf_la <- local_attributions(explain_titanic_rf, new_passanger)
rf_la#> contribution
#> Random Forest v7: intercept 0.325
#> Random Forest v7: age = 8 0.221
#> Random Forest v7: class = 1st 0.075
#> Random Forest v7: gender = male -0.049
#> Random Forest v7: fare = 72 -0.063
#> Random Forest v7: embarked = Southampton -0.020
#> Random Forest v7: sibsp = 0 -0.007
#> Random Forest v7: parch = 0 -0.028
#> Random Forest v7: prediction 0.455Plot attributions with D3
plotD3(rf_la)Calculate uncertainty for variable attributions
rf_la_un <- break_down_uncertainty(explain_titanic_rf, new_passanger,
path = "average")
plot(rf_la_un)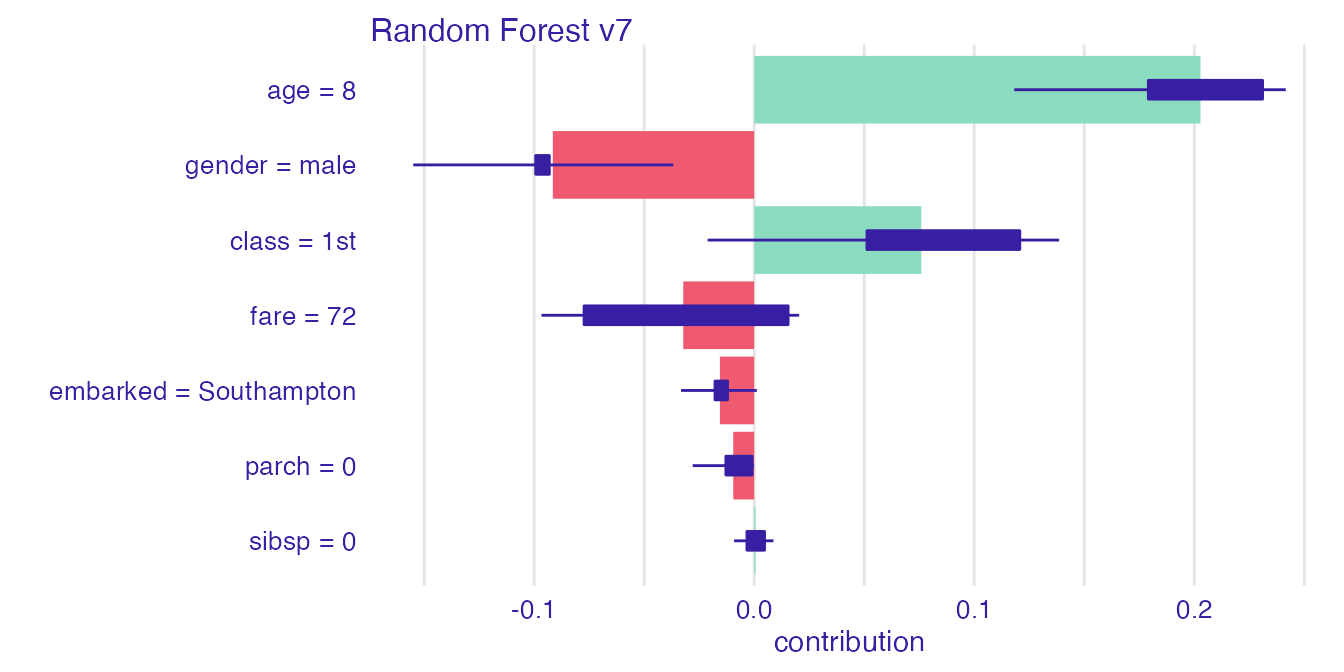
Show only top features
plotD3(rf_la, max_features = 3)