Dataset Level Variable Effect as Partial Dependency Profile or Accumulated Local Effects
Source:R/model_variable_effect.R
variable_effect.RdFrom DALEX version 1.0 this function calls the accumulated_dependence or
partial_dependence from the ingredients package.
Find information how to use this function here: https://ema.drwhy.ai/partialDependenceProfiles.html.
variable_effect(explainer, variables, ..., type = "partial_dependency")
variable_effect_partial_dependency(explainer, variables, ...)
variable_effect_accumulated_dependency(explainer, variables, ...)Arguments
- explainer
a model to be explained, preprocessed by the 'explain' function
- variables
character - names of variables to be explained
- ...
other parameters
- type
character - type of the response to be calculated. Currently following options are implemented: 'partial_dependency' for Partial Dependency and 'accumulated_dependency' for Accumulated Local Effects
Value
An object of the class 'aggregated_profiles_explainer'. It's a data frame with calculated average response.
References
Explanatory Model Analysis. Explore, Explain, and Examine Predictive Models. https://ema.drwhy.ai/
Examples
titanic_glm_model <- glm(survived~., data = titanic_imputed, family = "binomial")
explainer_glm <- explain(titanic_glm_model, data = titanic_imputed)
#> Preparation of a new explainer is initiated
#> -> model label : lm ( default )
#> -> data : 2207 rows 8 cols
#> -> target variable : not specified! ( WARNING )
#> -> predict function : yhat.glm will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package stats , ver. 4.2.3 , task classification ( default )
#> -> model_info : Model info detected classification task but 'y' is a NULL . ( WARNING )
#> -> model_info : By deafult classification tasks supports only numercical 'y' parameter.
#> -> model_info : Consider changing to numerical vector with 0 and 1 values.
#> -> model_info : Otherwise I will not be able to calculate residuals or loss function.
#> -> predicted values : numerical, min = 0.008128381 , mean = 0.3221568 , max = 0.9731431
#> -> residual function : difference between y and yhat ( default )
#> A new explainer has been created!
expl_glm <- variable_effect(explainer_glm, "fare", "partial_dependency")
#> Warning: 'variable_effect()' is deprecated. Use 'DALEX::model_profile()' instead.
#> Find examples and detailed introduction at: https://pbiecek.github.io/ema/
plot(expl_glm)
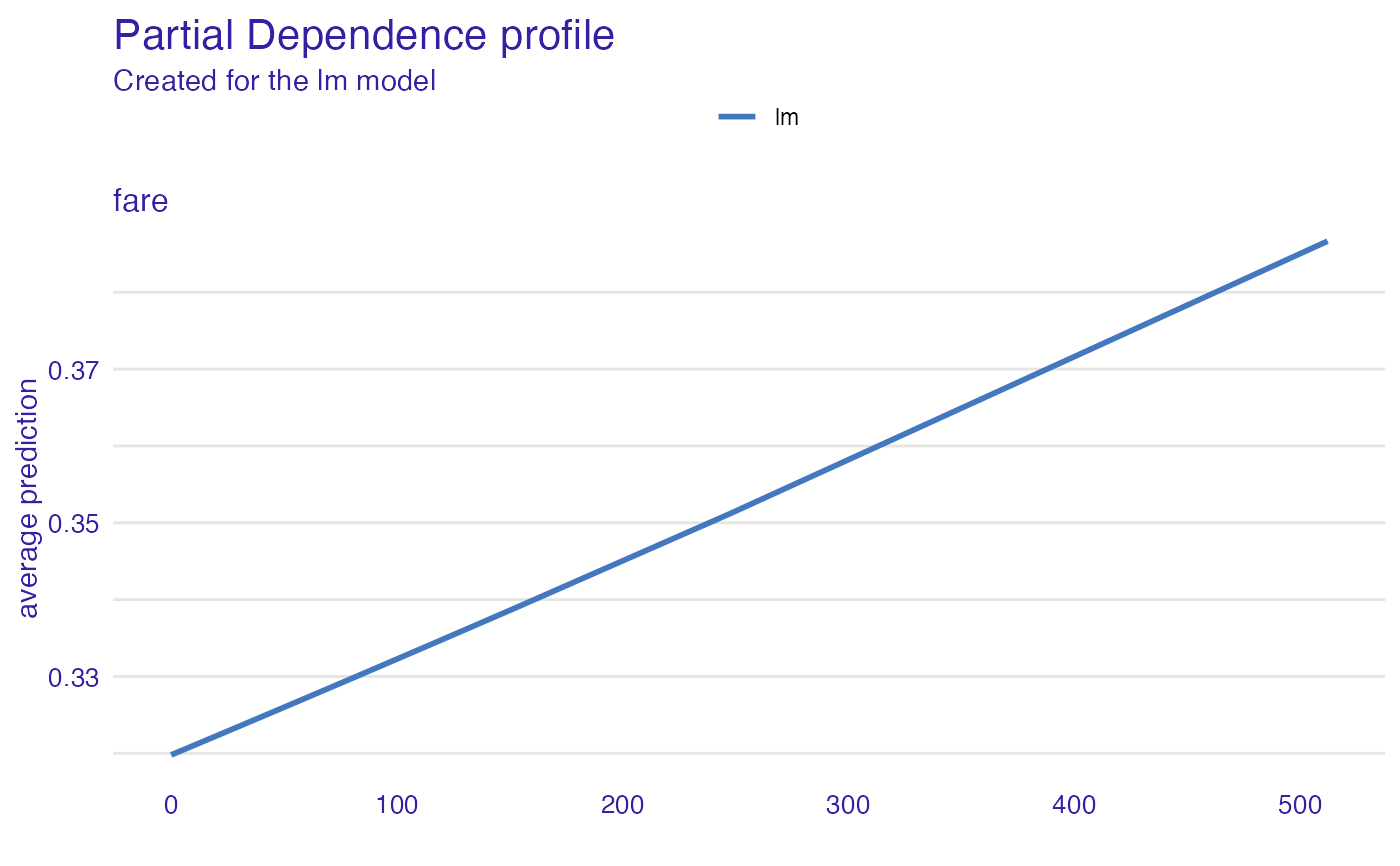 # \donttest{
library("ranger")
titanic_ranger_model <- ranger(survived~., data = titanic_imputed, num.trees = 50,
probability = TRUE)
explainer_ranger <- explain(titanic_ranger_model, data = titanic_imputed)
#> Preparation of a new explainer is initiated
#> -> model label : ranger ( default )
#> -> data : 2207 rows 8 cols
#> -> target variable : not specified! ( WARNING )
#> -> predict function : yhat.ranger will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package ranger , ver. 0.14.1 , task classification ( default )
#> -> model_info : Model info detected classification task but 'y' is a NULL . ( WARNING )
#> -> model_info : By deafult classification tasks supports only numercical 'y' parameter.
#> -> model_info : Consider changing to numerical vector with 0 and 1 values.
#> -> model_info : Otherwise I will not be able to calculate residuals or loss function.
#> -> predicted values : numerical, min = 0.01281399 , mean = 0.3200535 , max = 0.9984948
#> -> residual function : difference between y and yhat ( default )
#> A new explainer has been created!
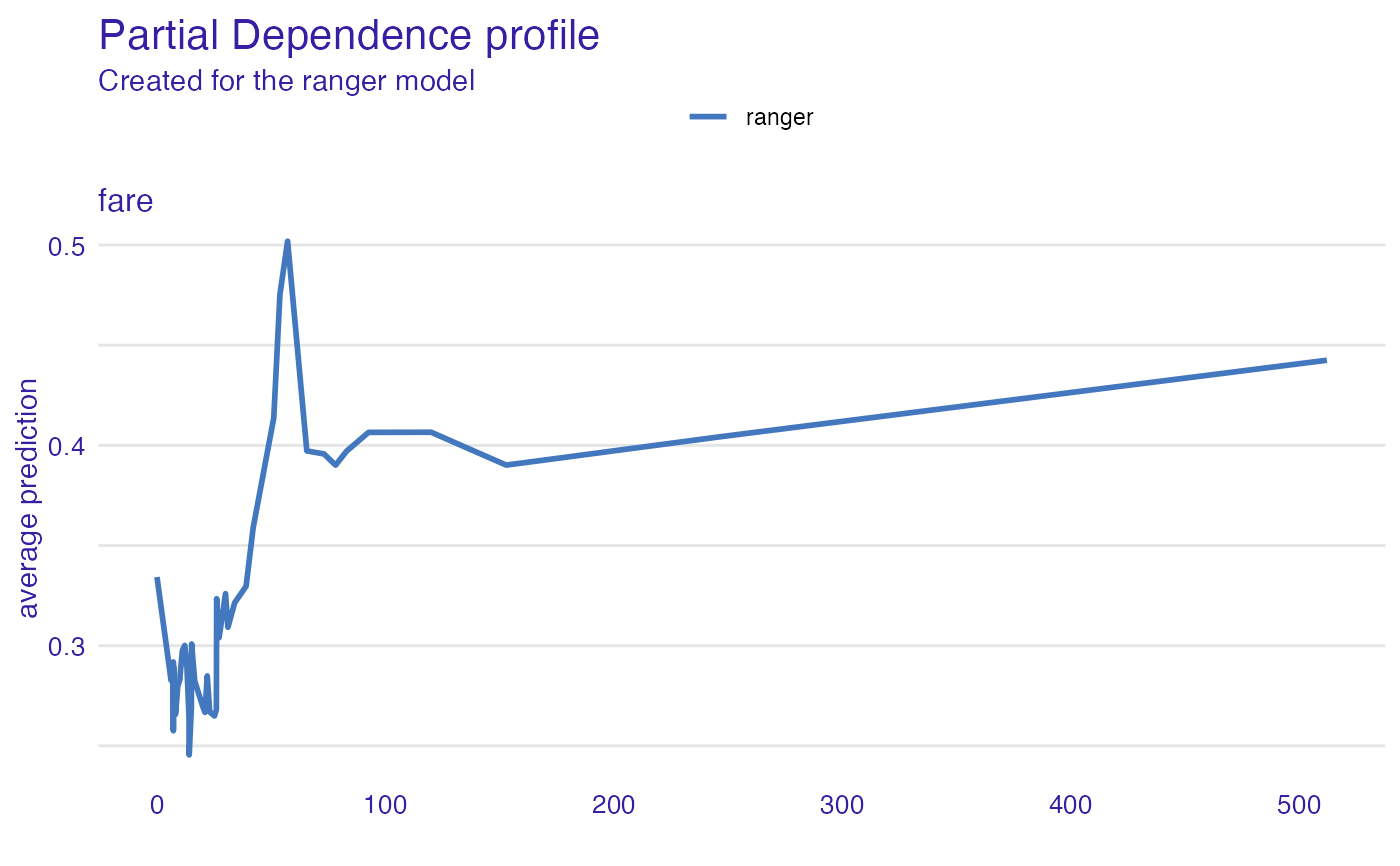
expl_ranger <- variable_effect(explainer_ranger, variables = "fare",
type = "partial_dependency")
plot(expl_ranger)
# \donttest{
library("ranger")
titanic_ranger_model <- ranger(survived~., data = titanic_imputed, num.trees = 50,
probability = TRUE)
explainer_ranger <- explain(titanic_ranger_model, data = titanic_imputed)
#> Preparation of a new explainer is initiated
#> -> model label : ranger ( default )
#> -> data : 2207 rows 8 cols
#> -> target variable : not specified! ( WARNING )
#> -> predict function : yhat.ranger will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package ranger , ver. 0.14.1 , task classification ( default )
#> -> model_info : Model info detected classification task but 'y' is a NULL . ( WARNING )
#> -> model_info : By deafult classification tasks supports only numercical 'y' parameter.
#> -> model_info : Consider changing to numerical vector with 0 and 1 values.
#> -> model_info : Otherwise I will not be able to calculate residuals or loss function.
#> -> predicted values : numerical, min = 0.01281399 , mean = 0.3200535 , max = 0.9984948
#> -> residual function : difference between y and yhat ( default )
#> A new explainer has been created!
expl_ranger <- variable_effect(explainer_ranger, variables = "fare",
type = "partial_dependency")
plot(expl_ranger)
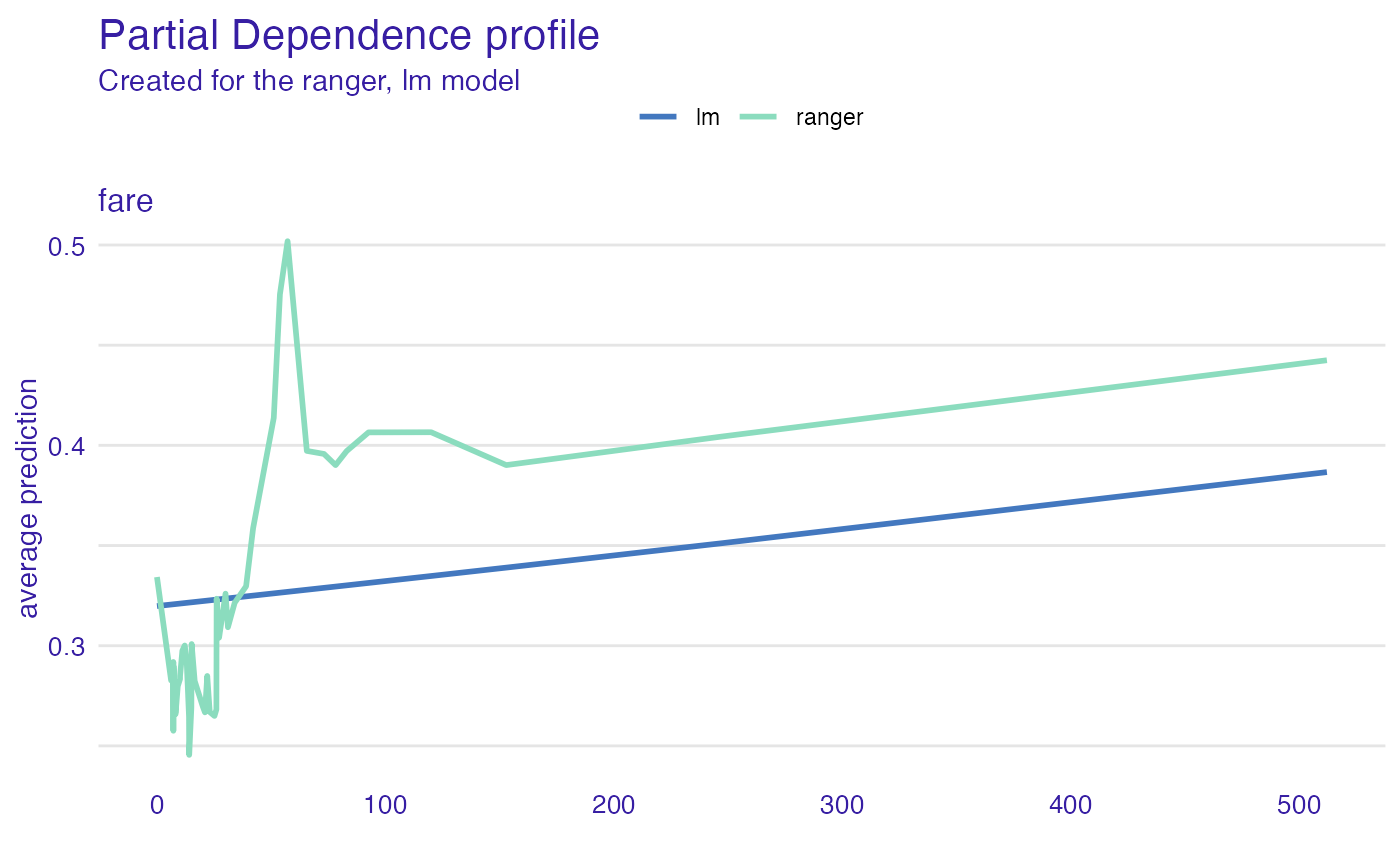
 plot(expl_ranger, expl_glm)
plot(expl_ranger, expl_glm)
 # Example for factor variable (with factorMerger)
expl_ranger_factor <- variable_effect(explainer_ranger, variables = "class")
#> 'variable_type' changed to 'categorical' due to lack of numerical variables.
plot(expl_ranger_factor)
# Example for factor variable (with factorMerger)
expl_ranger_factor <- variable_effect(explainer_ranger, variables = "class")
#> 'variable_type' changed to 'categorical' due to lack of numerical variables.
plot(expl_ranger_factor)
 # }
# }