Accumulated Local Effects Profiles aka ALEPlots
Source:R/accumulated_dependence.R
accumulated_dependence.RdAccumulated Local Effects Profiles accumulate local changes in Ceteris Paribus Profiles.
Function accumulated_dependence calls ceteris_paribus and then aggregate_profiles.
accumulated_dependence(x, ...)
# S3 method for explainer
accumulated_dependence(
x,
variables = NULL,
N = 500,
variable_splits = NULL,
grid_points = 101,
...,
variable_type = "numerical"
)
# S3 method for default
accumulated_dependence(
x,
data,
predict_function = predict,
label = class(x)[1],
variables = NULL,
N = 500,
variable_splits = NULL,
grid_points = 101,
...,
variable_type = "numerical"
)
# S3 method for ceteris_paribus_explainer
accumulated_dependence(x, ..., variables = NULL)
accumulated_dependency(x, ...)Arguments
- x
an explainer created with function
DALEX::explain(), an object of the classceteris_paribus_explaineror a model to be explained.- ...
other parameters
- variables
names of variables for which profiles shall be calculated. Will be passed to
calculate_variable_split. IfNULLthen all variables from the validation data will be used.- N
number of observations used for calculation of partial dependence profiles. By default,
500observations will be chosen randomly.- variable_splits
named list of splits for variables, in most cases created with
calculate_variable_split. IfNULLthen it will be calculated based on validation data avaliable in theexplainer.- grid_points
number of points for profile. Will be passed to
calculate_variable_split.- variable_type
a character. If
"numerical"then only numerical variables will be calculated. If"categorical"then only categorical variables will be calculated.- data
validation dataset Will be extracted from
xif it's an explainer NOTE: It is best when target variable is not present in thedata- predict_function
predict function Will be extracted from
xif it's an explainer- label
name of the model. By default it's extracted from the
classattribute of the model
Value
an object of the class aggregated_profiles_explainer
Details
Find more detailes in the Accumulated Local Dependence Chapter.
References
ALEPlot: Accumulated Local Effects (ALE) Plots and Partial Dependence (PD) Plots https://cran.r-project.org/package=ALEPlot, Explanatory Model Analysis. Explore, Explain, and Examine Predictive Models. https://ema.drwhy.ai/
Examples
library("DALEX")
#> Welcome to DALEX (version: 2.4.2).
#> Find examples and detailed introduction at: http://ema.drwhy.ai/
#> Additional features will be available after installation of: ggpubr.
#> Use 'install_dependencies()' to get all suggested dependencies
#>
#> Attaching package: ‘DALEX’
#> The following object is masked from ‘package:ingredients’:
#>
#> feature_importance
library("ingredients")
model_titanic_glm <- glm(survived ~ gender + age + fare,
data = titanic_imputed, family = "binomial")
explain_titanic_glm <- explain(model_titanic_glm,
data = titanic_imputed[,-8],
y = titanic_imputed[,8],
verbose = FALSE)
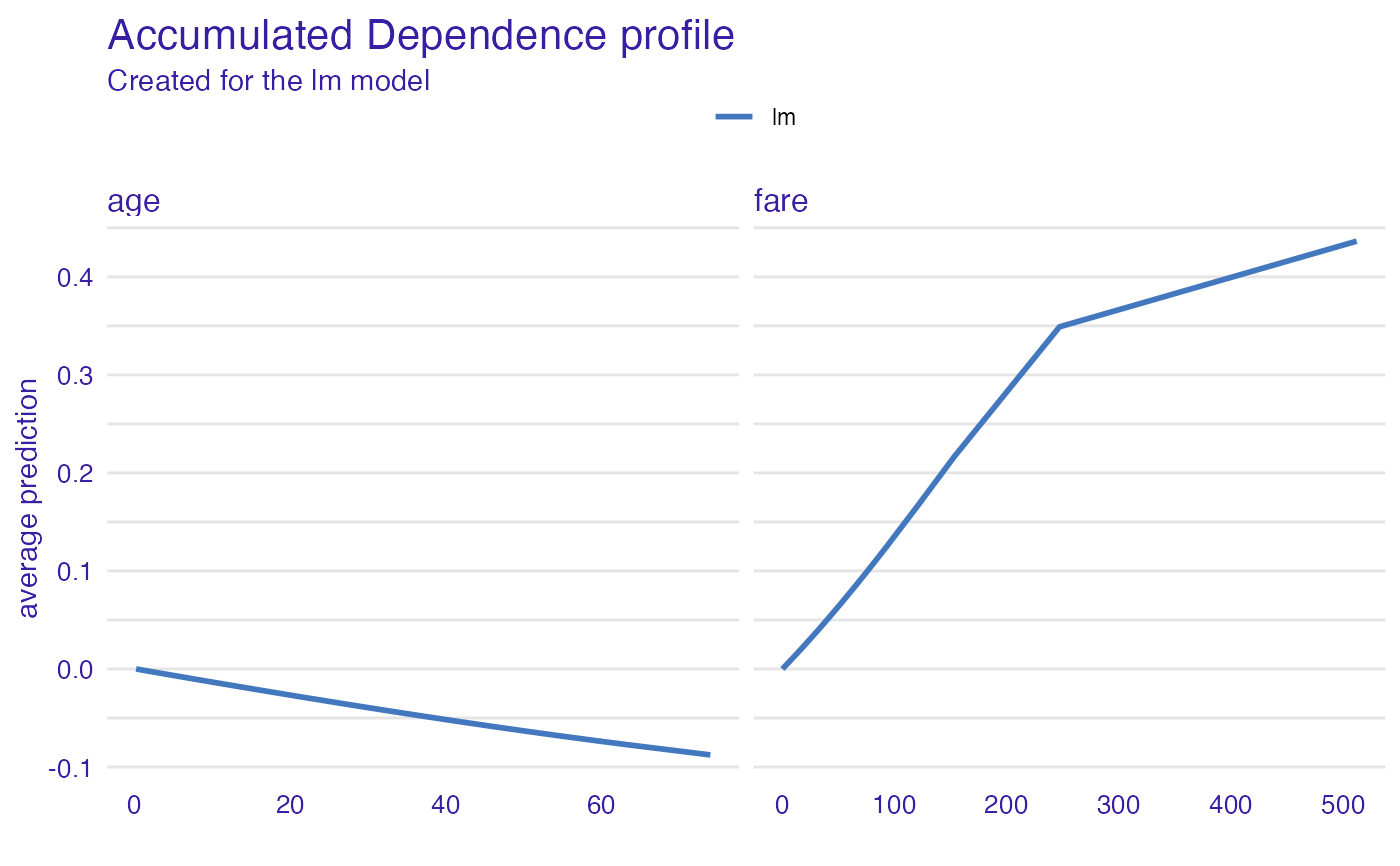
adp_glm <- accumulated_dependence(explain_titanic_glm,
N = 25, variables = c("age", "fare"))
head(adp_glm)
#> Top profiles :
#> _vname_ _label_ _x_ _yhat_ _ids_
#> 1 age lm 0.1666667 0.000000000 0
#> 2 age lm 2.0000000 -0.002538769 0
#> 3 age lm 4.0000000 -0.005291217 0
#> 4 age lm 7.0000000 -0.009385779 0
#> 5 age lm 9.0000000 -0.012095000 0
#> 6 age lm 13.0000000 -0.017457612 0
plot(adp_glm)
 # \donttest{
library("ranger")
model_titanic_rf <- ranger(survived ~., data = titanic_imputed, probability = TRUE)
explain_titanic_rf <- explain(model_titanic_rf,
data = titanic_imputed[,-8],
y = titanic_imputed[,8],
label = "ranger forest",
verbose = FALSE)
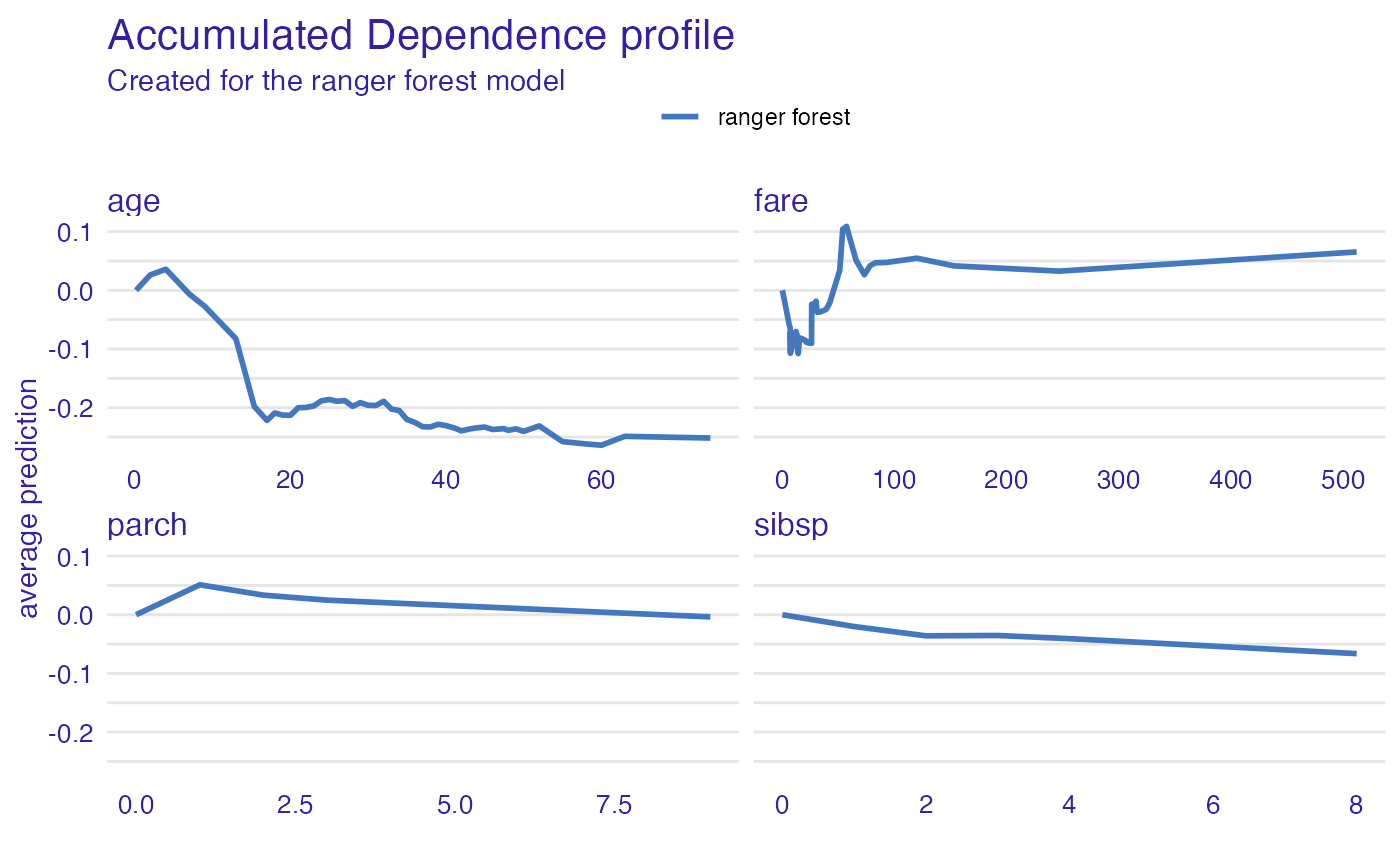
adp_rf <- accumulated_dependence(explain_titanic_rf, N = 200, variable_type = "numerical")
plot(adp_rf)
# \donttest{
library("ranger")
model_titanic_rf <- ranger(survived ~., data = titanic_imputed, probability = TRUE)
explain_titanic_rf <- explain(model_titanic_rf,
data = titanic_imputed[,-8],
y = titanic_imputed[,8],
label = "ranger forest",
verbose = FALSE)
adp_rf <- accumulated_dependence(explain_titanic_rf, N = 200, variable_type = "numerical")
plot(adp_rf)
 adp_rf <- accumulated_dependence(explain_titanic_rf, N = 200, variable_type = "categorical")
plotD3(adp_rf, label_margin = 80, scale_plot = TRUE)
# }
adp_rf <- accumulated_dependence(explain_titanic_rf, N = 200, variable_type = "categorical")
plotD3(adp_rf, label_margin = 80, scale_plot = TRUE)
# }