LIME kernel that treats all observations as equally similar to the observation of interest.
Source:R/kernels.R
identity_kernel.RdKernels are meant to be used as an argument to individual_surrogate_model function. Other custom functions can be used. Such functions take two vectors and return a single number.
identity_kernel(explained_instance, simulated_instance)
Arguments
| explained_instance | explained instance |
|---|---|
| simulated_instance | new observation |
Value
numeric
Examples
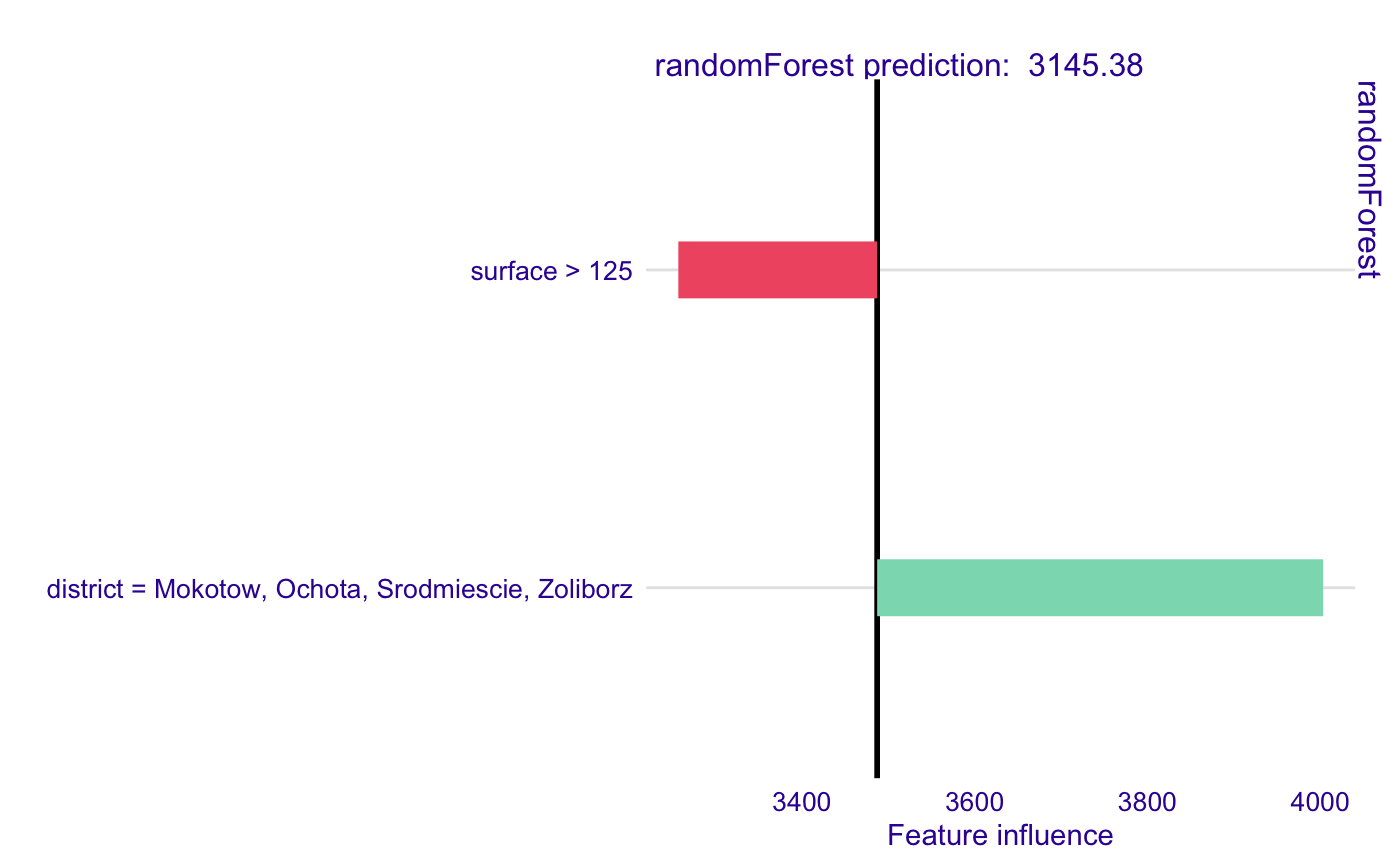
library(DALEX) library(randomForest) library(localModel) data('apartments') mrf <- randomForest(m2.price ~., data = apartments, ntree = 50) explainer <- explain(model = mrf, data = apartments[, -1])#> Preparation of a new explainer is initiated #> -> model label : randomForest ( default ) #> -> data : 1000 rows 5 cols #> -> target variable : not specified! ( WARNING ) #> -> predict function : yhat.randomForest will be used ( default ) #> -> predicted values : numerical, min = 1917.419 , mean = 3486.937 , max = 5842.92 #> -> model_info : package randomForest , ver. 4.6.14 , task regression ( default ) #> -> residual function : difference between y and yhat ( default ) #> A new explainer has been created!model_lok <- individual_surrogate_model(explainer, apartments[5, -1], size = 500, seed = 17, kernel = identity_kernel) # In this case each simulated observation has equal weight # when explanation model (LASSO) is fitted. model_lok#> estimated variable original_variable #> 1 3486.9367 (Model mean) #> 2 3245.5658 (Intercept) #> 3 -230.1591 surface > 125 surface #> 4 516.2736 district = Mokotow, Ochota, Srodmiescie, Zoliborz district #> dev_ratio response predicted_value model #> 1 0.5241099 3145.381 randomForest #> 2 0.5241099 3145.381 randomForest #> 3 0.5241099 3145.381 randomForest #> 4 0.5241099 3145.381 randomForestplot(model_lok)