LIME-like explanations based on Ceteris Paribus curves
Source:R/local_surrogate.R
individual_surrogate_model.RdThis function fits a LIME-type explanation of a single prediction. Interpretable binary features that describe the local impact of features on the prediction are created based on Ceteris Paribus Profiles. Thend, a new dataset of similar observations is created and black box model predictions (scores in case of classification) are calculated for this dataset and LASSO regression model is fitted to them. This way, explanations are simplified and include only the most important features. More details about the methodology can be found in the vignettes.
individual_surrogate_model( x, new_observation, size, seed = NULL, kernel = identity_kernel, sampling = "uniform", ... )
Arguments
| x | an explainer created with the function DALEX::explain(). |
|---|---|
| new_observation | an observation to be explained. Columns in should correspond to columns in the data argument to x. |
| size | number of similar observation to be sampled. |
| seed | If not NULL, seed will be set to this value for reproducibility. |
| kernel | Kernel function which will be used to weight simulated observations. |
| sampling | Parameter that controls sampling while creating new observations. |
| ... | Additional arguments that will be passed to ingredients::ceteris_paribus. |
Value
data.frame of class local_surrogate_explainer
Examples
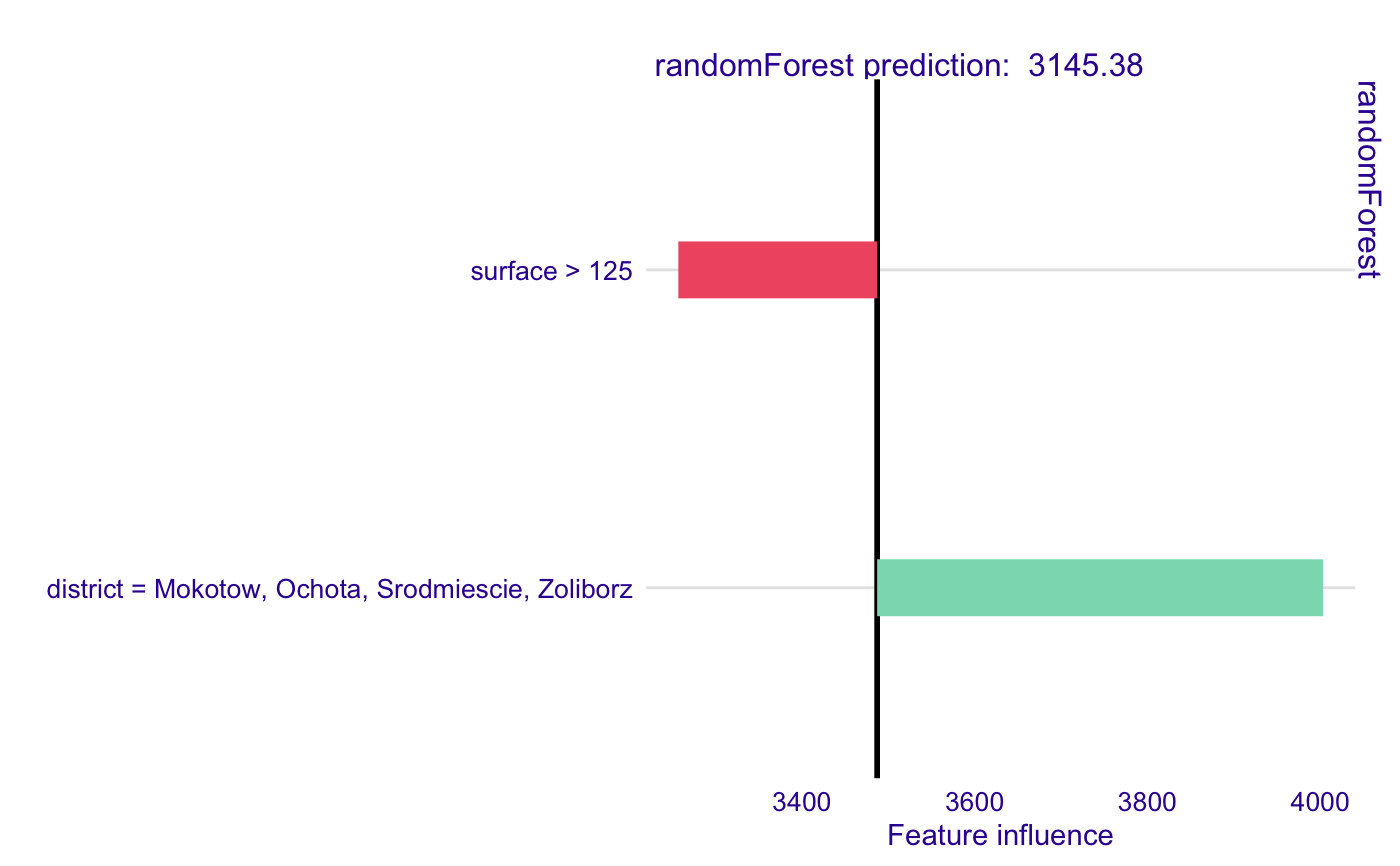
# Example based on apartments data from DALEX package. library(DALEX) library(randomForest) library(localModel) data('apartments') mrf <- randomForest(m2.price ~., data = apartments, ntree = 50) explainer <- explain(model = mrf, data = apartments[, -1])#> Preparation of a new explainer is initiated #> -> model label : randomForest ( default ) #> -> data : 1000 rows 5 cols #> -> target variable : not specified! ( WARNING ) #> -> predict function : yhat.randomForest will be used ( default ) #> -> predicted values : numerical, min = 1917.419 , mean = 3486.937 , max = 5842.92 #> -> model_info : package randomForest , ver. 4.6.14 , task regression ( default ) #> -> residual function : difference between y and yhat ( default ) #> A new explainer has been created!model_lok <- individual_surrogate_model(explainer, apartments[5, -1], size = 500, seed = 17) model_lok#> estimated variable original_variable #> 1 3486.9367 (Model mean) #> 2 3245.5658 (Intercept) #> 3 -230.1591 surface > 125 surface #> 4 516.2736 district = Mokotow, Ochota, Srodmiescie, Zoliborz district #> dev_ratio response predicted_value model #> 1 0.5241099 3145.381 randomForest #> 2 0.5241099 3145.381 randomForest #> 3 0.5241099 3145.381 randomForest #> 4 0.5241099 3145.381 randomForestplot(model_lok)