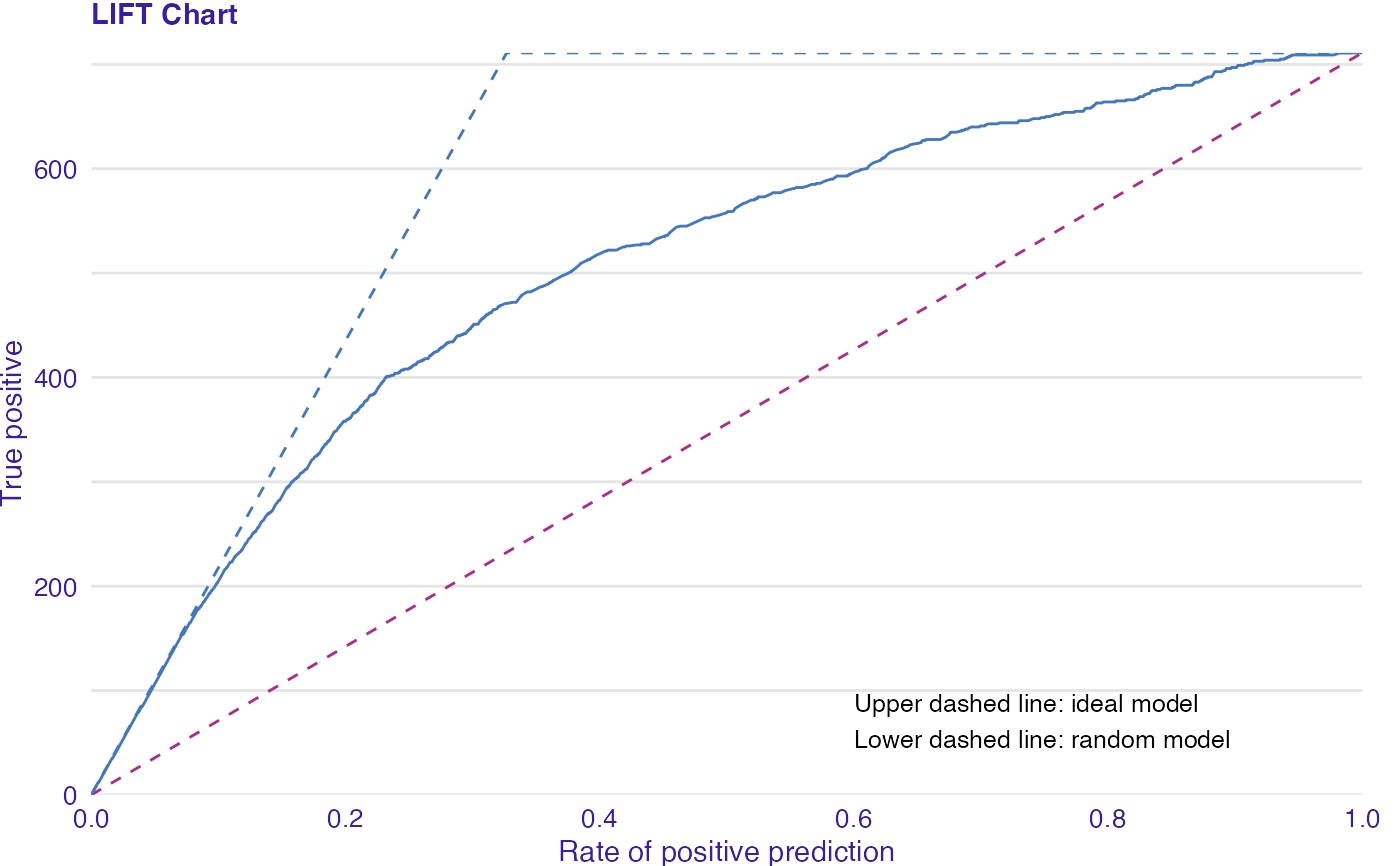
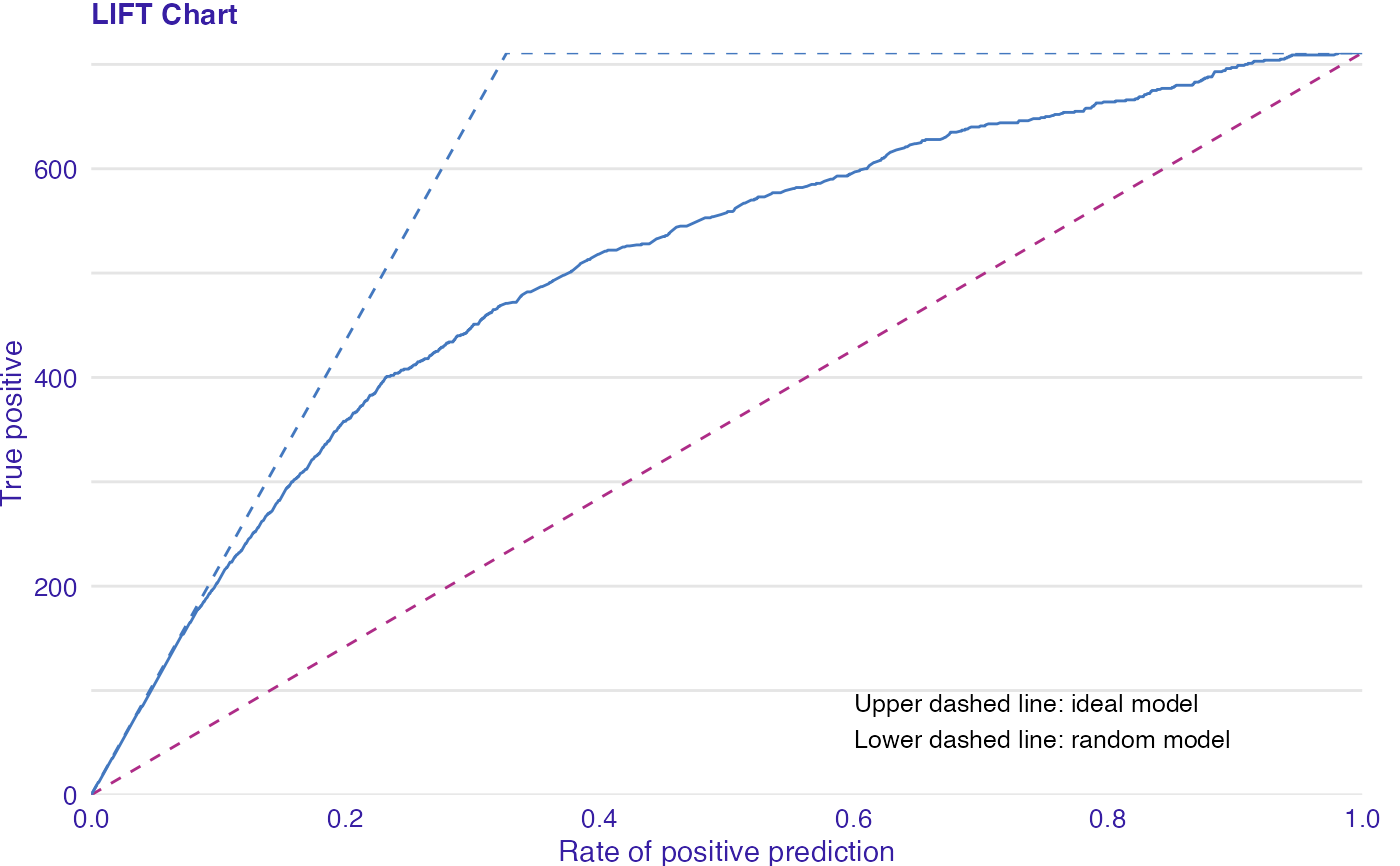
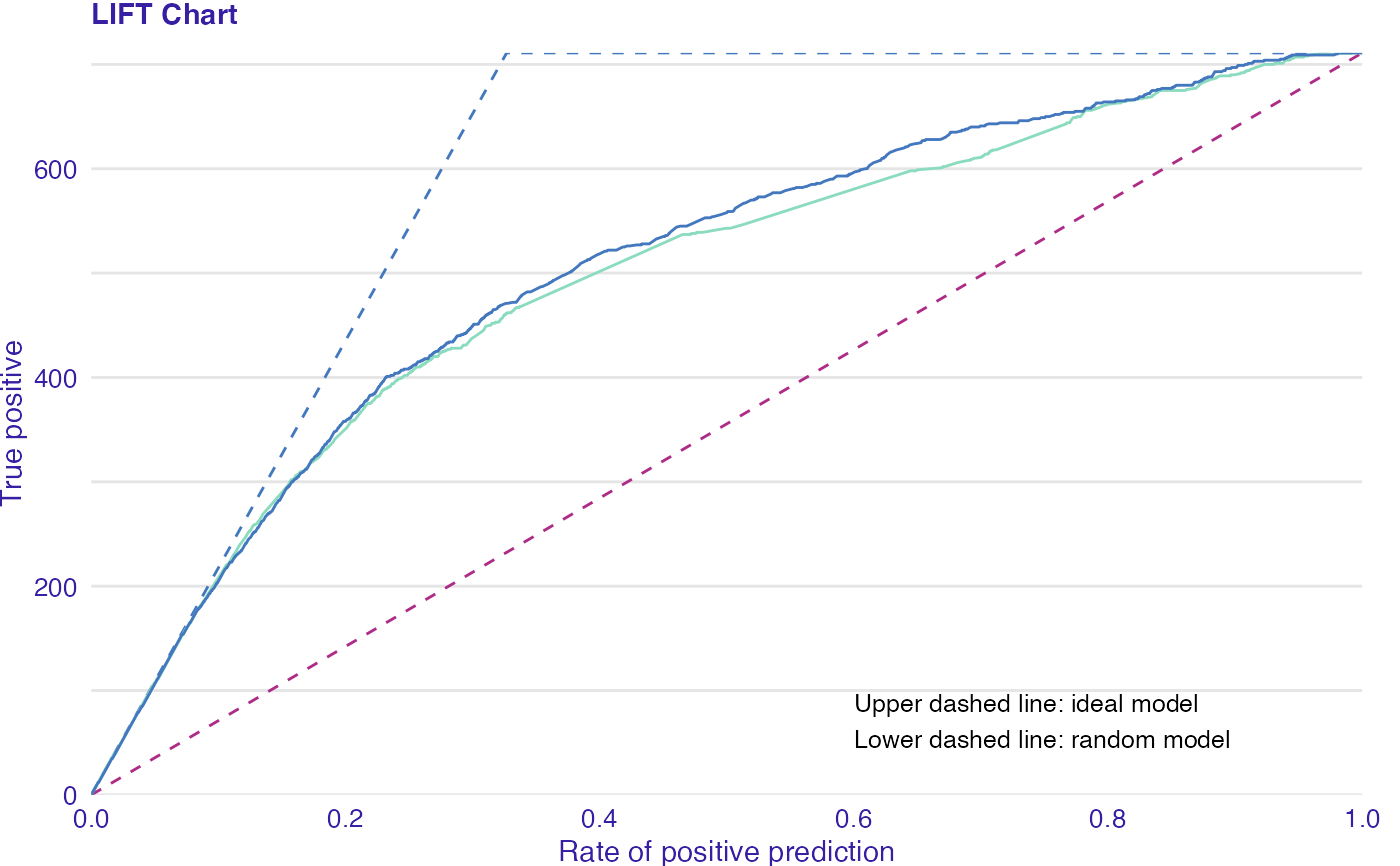
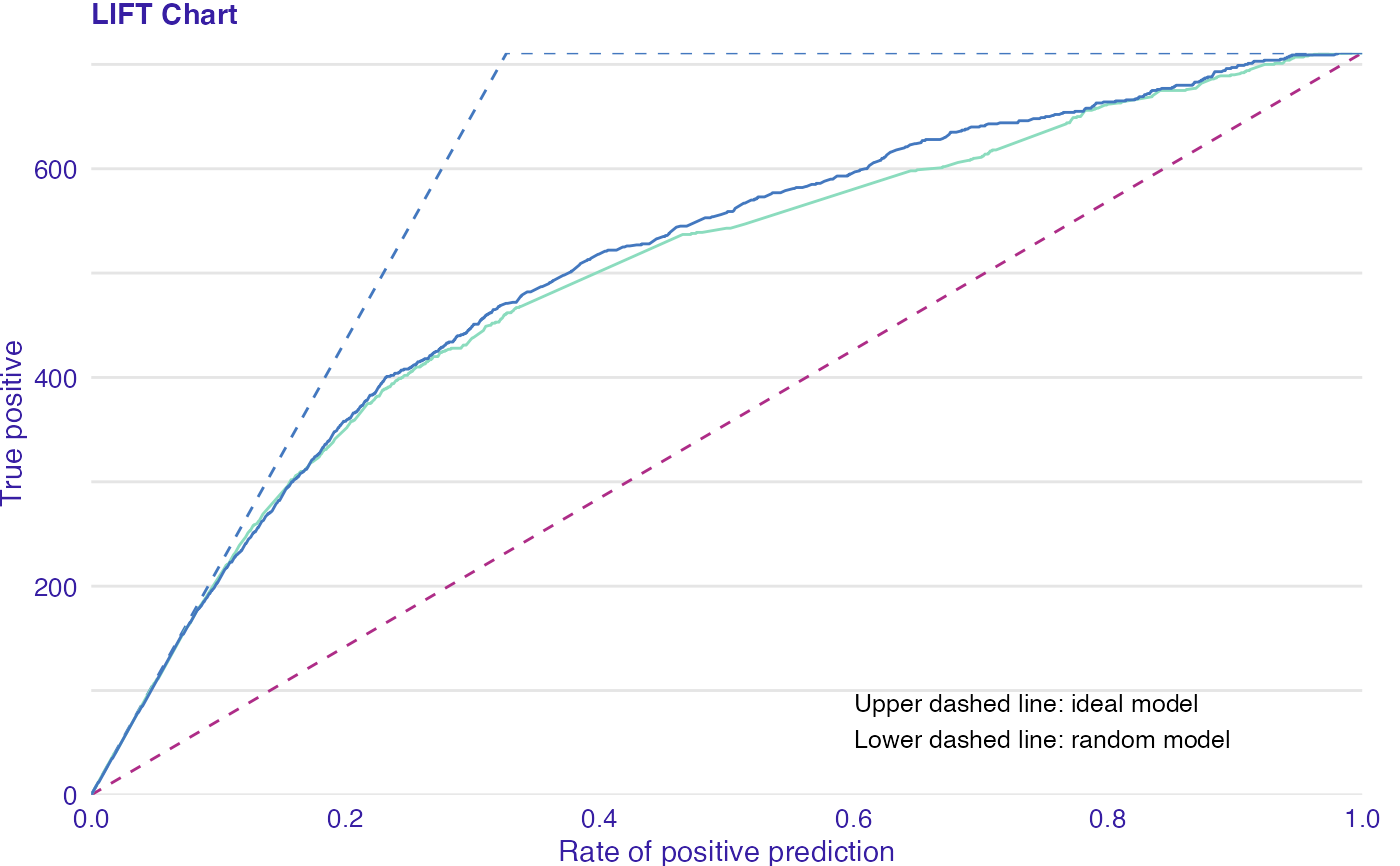
LIFT is a plot of the rate of positive prediction against true positive rate for the different thresholds. It is useful for measuring and comparing the accuracy of the classificators.
plot_lift(object, ..., zeros = TRUE) plotLIFT(object, ...)
Arguments
| object | An object of class |
|---|---|
| ... | Other |
| zeros | Logical. It makes the lines start from the |
Value
A ggplot object.
See also
Examples
data(titanic_imputed, package = "DALEX") # fit a model model_glm <- glm(survived ~ ., family = binomial, data = titanic_imputed) glm_audit <- audit(model_glm, data = titanic_imputed, y = titanic_imputed$survived)#> Preparation of a new explainer is initiated #> -> model label : lm ( default ) #> -> data : 2207 rows 8 cols #> -> target variable : 2207 values #> -> predict function : yhat.glm will be used ( default ) #> -> predicted values : No value for predict function target column. ( default ) #> -> model_info : package stats , ver. 4.1.1 , task classification ( default ) #> -> predicted values : numerical, min = 0.008128381 , mean = 0.3221568 , max = 0.9731431 #> -> residual function : difference between y and yhat ( default ) #> -> residuals : numerical, min = -0.9628583 , mean = -2.569729e-10 , max = 0.9663346 #> A new explainer has been created!# validate a model with auditor eva_glm <- model_evaluation(glm_audit) # plot results plot_lift(eva_glm)model_glm_2 <- glm(survived ~ .-age, family = binomial, data = titanic_imputed) glm_audit_2 <- audit(model_glm_2, data = titanic_imputed, y = titanic_imputed$survived, label = "glm2")#> Preparation of a new explainer is initiated #> -> model label : glm2 #> -> data : 2207 rows 8 cols #> -> target variable : 2207 values #> -> predict function : yhat.glm will be used ( default ) #> -> predicted values : No value for predict function target column. ( default ) #> -> model_info : package stats , ver. 4.1.1 , task classification ( default ) #> -> predicted values : numerical, min = 0.01618585 , mean = 0.3221568 , max = 0.9628841 #> -> residual function : difference between y and yhat ( default ) #> -> residuals : numerical, min = -0.9354237 , mean = -1.981381e-10 , max = 0.9654417 #> A new explainer has been created!