This function calculates martingale and deviance residuals.
model_diagnostics(explainer)
# S3 method for surv_explainer
model_diagnostics(explainer)Arguments
- explainer
an explainer object - model preprocessed by the
explain()function
Value
An object of class c("model_diagnostics_survival"). It's a list with the explanations in the result element.
Examples
# \donttest{
library(survival)
library(survex)
cph <- coxph(Surv(time, status) ~ ., data = veteran, model = TRUE, x = TRUE, y = TRUE)
rsf_ranger <- ranger::ranger(Surv(time, status) ~ .,
data = veteran,
respect.unordered.factors = TRUE,
num.trees = 100,
mtry = 3,
max.depth = 5
)
cph_exp <- explain(cph)
#> Preparation of a new explainer is initiated
#> -> model label : coxph ( default )
#> -> data : 137 rows 6 cols ( extracted from the model )
#> -> target variable : 137 values ( 128 events and 9 censored , censoring rate = 0.066 ) ( extracted from the model )
#> -> times : 50 unique time points , min = 1.5 , median survival time = 80 , max = 999
#> -> times : ( generated from y as uniformly distributed survival quantiles based on Kaplan-Meier estimator )
#> -> predict function : predict.coxph with type = 'risk' will be used ( default )
#> -> predict survival function : predictSurvProb.coxph will be used ( default )
#> -> predict cumulative hazard function : -log(predict_survival_function) will be used ( default )
#> -> model_info : package survival , ver. 3.7.0 , task survival ( default )
#> A new explainer has been created!
rsf_ranger_exp <- explain(rsf_ranger,
data = veteran[, -c(3, 4)],
y = Surv(veteran$time, veteran$status)
)
#> Preparation of a new explainer is initiated
#> -> model label : ranger ( default )
#> -> data : 137 rows 6 cols
#> -> target variable : 137 values ( 128 events and 9 censored )
#> -> times : 50 unique time points , min = 1.5 , median survival time = 80 , max = 999
#> -> times : ( generated from y as uniformly distributed survival quantiles based on Kaplan-Meier estimator )
#> -> predict function : sum over the predict_cumulative_hazard_function will be used ( default )
#> -> predict survival function : stepfun based on predict.ranger()$survival will be used ( default )
#> -> predict cumulative hazard function : stepfun based on predict.ranger()$chf will be used ( default )
#> -> model_info : package ranger , ver. 0.16.0 , task survival ( default )
#> A new explainer has been created!
cph_residuals <- model_diagnostics(cph_exp)
rsf_residuals <- model_diagnostics(rsf_ranger_exp)
head(cph_residuals$result)
#> time status cox_snell_residuals martingale_residuals deviance_residuals label
#> 1 72 1 0.2547163 0.7452837 1.1156354 coxph
#> 2 411 1 1.4900714 -0.4900714 -0.4271940 coxph
#> 3 228 1 1.1846769 -0.1846769 -0.1743951 coxph
#> 4 126 1 0.5863747 0.4136253 0.4902469 coxph
#> 5 118 1 0.3919334 0.6080666 0.8106747 coxph
#> 6 10 1 0.1382539 0.8617461 1.4946018 coxph
#> trt celltype karno diagtime age prior
#> 1 1 squamous 60 7 69 0
#> 2 1 squamous 70 5 64 10
#> 3 1 squamous 60 3 38 0
#> 4 1 squamous 60 9 63 10
#> 5 1 squamous 70 11 65 10
#> 6 1 squamous 20 5 49 0
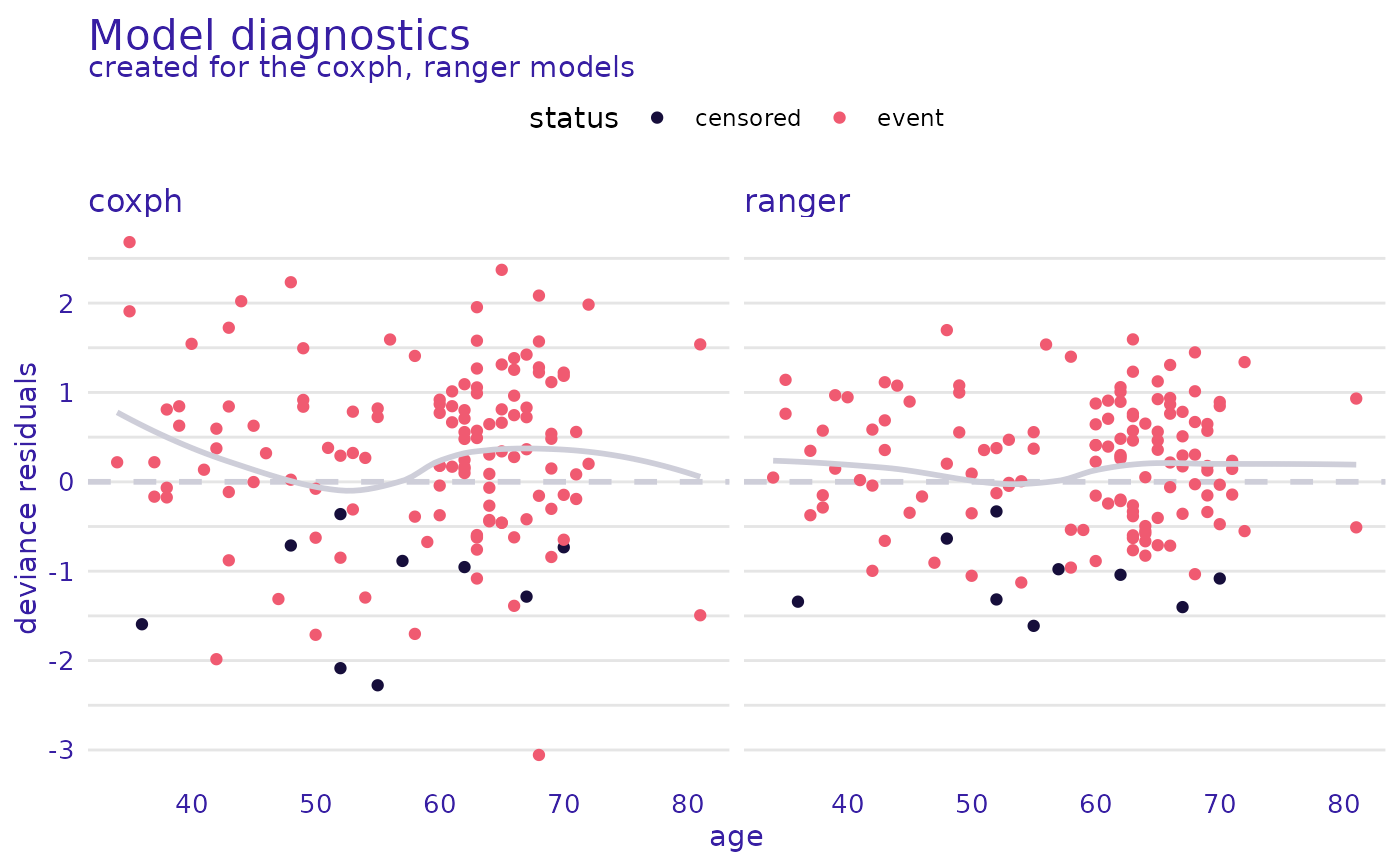
plot(cph_residuals, rsf_residuals, xvariable = "age")
#> `geom_smooth()` using method = 'loess' and formula = 'y ~ x'
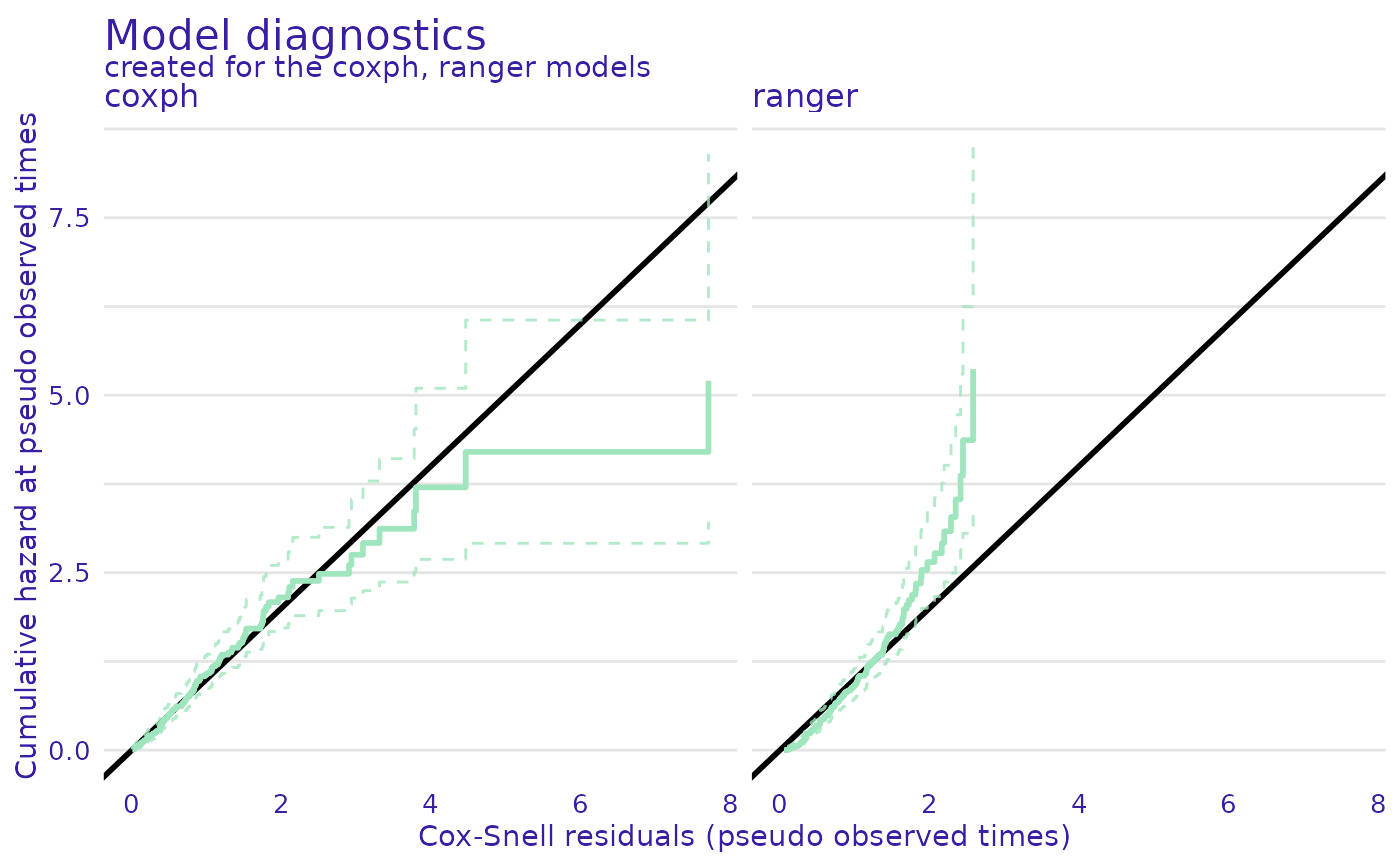
 plot(cph_residuals, rsf_residuals, plot_type = "Cox-Snell")
plot(cph_residuals, rsf_residuals, plot_type = "Cox-Snell")
 # }
# }