Calculates the average loss of a model on a given dataset,
optionally grouped by a variable. Use plot() to visualize the results.
average_loss(object, ...)
# Default S3 method
average_loss(
object,
X,
y,
pred_fun = stats::predict,
loss = "squared_error",
agg_cols = FALSE,
BY = NULL,
by_size = 4L,
w = NULL,
...
)
# S3 method for class 'ranger'
average_loss(
object,
X,
y,
pred_fun = function(m, X, ...) stats::predict(m, X, ...)$predictions,
loss = "squared_error",
agg_cols = FALSE,
BY = NULL,
by_size = 4L,
w = NULL,
...
)
# S3 method for class 'explainer'
average_loss(
object,
X = object[["data"]],
y = object[["y"]],
pred_fun = object[["predict_function"]],
loss = "squared_error",
agg_cols = FALSE,
BY = NULL,
by_size = 4L,
w = object[["weights"]],
...
)Arguments
- object
Fitted model object.
- ...
Additional arguments passed to
pred_fun(object, X, ...), for instancetype = "response"in aglm()model, orreshape = TRUEin a multiclass XGBoost model.- X
A data.frame or matrix serving as background dataset.
- y
Vector/matrix of the response, or the corresponding column names in
X.- pred_fun
Prediction function of the form
function(object, X, ...), providing \(K \ge 1\) predictions per row. Its first argument represents the modelobject, its second argument a data structure likeX. Additional arguments (such astype = "response"in a GLM, orreshape = TRUEin a multiclass XGBoost model) can be passed via.... The default,stats::predict(), will work in most cases.- loss
One of "squared_error", "logloss", "mlogloss", "poisson", "gamma", or "absolute_error". Alternatively, a loss function can be provided that turns observed and predicted values into a numeric vector or matrix of unit losses of the same length as
X. For "mlogloss", the responseycan either be a dummy matrix or a discrete vector. The latter case is handled via a fast version ofmodel.matrix(~ as.factor(y) + 0). For "squared_error", the response can be a factor with levels in column order of the predictions. In this case, squared error is evaluated for each one-hot-encoded column.- agg_cols
Should multivariate losses be summed up? Default is
FALSE. In combination with the squared error loss,agg_cols = TRUEgives the Brier score for (probabilistic) classification.- BY
Optional grouping vector or column name. Numeric
BYvariables with more thanby_sizedisjoint values will be binned intoby_sizequantile groups of similar size.- by_size
Numeric
BYvariables with more thanby_sizeunique values will be binned into quantile groups. Only relevant ifBYis notNULL.- w
Optional vector of case weights. Can also be a column name of
X.
Value
An object of class "hstats_matrix" containing these elements:
M: Matrix of statistics (one column per prediction dimension), orNULL.SE: Matrix with standard errors ofM, orNULL. Multiply withsqrt(m_rep)to get standard deviations instead. Currently, supported only forperm_importance().m_rep: The number of repetitions behind standard errorsSE, orNULL. Currently, supported only forperm_importance().statistic: Name of the function that generated the statistic.description: Description of the statistic.
Methods (by class)
average_loss(default): Default method.average_loss(ranger): Method for "ranger" models.average_loss(explainer): Method for DALEX "explainer".
Losses
The default loss is the "squared_error". Other choices:
"absolute_error": The absolute error is the loss corresponding to median regression.
"poisson": Unit Poisson deviance, i.e., the loss function used in Poisson regression. Actual values
yand predictions must be non-negative."gamma": Unit gamma deviance, i.e., the loss function of Gamma regression. Actual values
yand predictions must be positive."logloss": The Log Loss is the loss function used in logistic regression, and the top choice in probabilistic binary classification. Responses
yand predictions must be between 0 and 1. Predictions represent probabilities of having a "1"."mlogloss": Multi-Log-Loss is the natural loss function in probabilistic multi-class situations. If there are K classes and n observations, the predictions form a (n x K) matrix of probabilities (with row-sums 1). The observed values
yare either passed as (n x K) dummy matrix, or as discrete vector with corresponding levels. The latter case is turned into a dummy matrix by a fast version ofmodel.matrix(~ as.factor(y) + 0).A function with signature
f(actual, predicted), returning a numeric vector or matrix of the same length as the input.
Examples
# MODEL 1: Linear regression
fit <- lm(Sepal.Length ~ ., data = iris)
average_loss(fit, X = iris, y = "Sepal.Length")
#> Average loss
#> [1] 0.09037657
average_loss(fit, X = iris, y = iris$Sepal.Length, BY = iris$Sepal.Width)
#> Average loss
#> [2,2.8] (2.8,3] (3,3.3] (3.3,4.4]
#> 0.08934441 0.09847409 0.09612552 0.07914772
average_loss(fit, X = iris, y = "Sepal.Length", BY = "Sepal.Width")
#> Average loss
#> [2,2.8] (2.8,3] (3,3.3] (3.3,4.4]
#> 0.08934441 0.09847409 0.09612552 0.07914772
# MODEL 2: Multi-response linear regression
fit <- lm(as.matrix(iris[, 1:2]) ~ Petal.Length + Petal.Width + Species, data = iris)
average_loss(fit, X = iris, y = iris[, 1:2])
#> Average loss
#> Sepal.Length Sepal.Width
#> 0.11120993 0.08472089
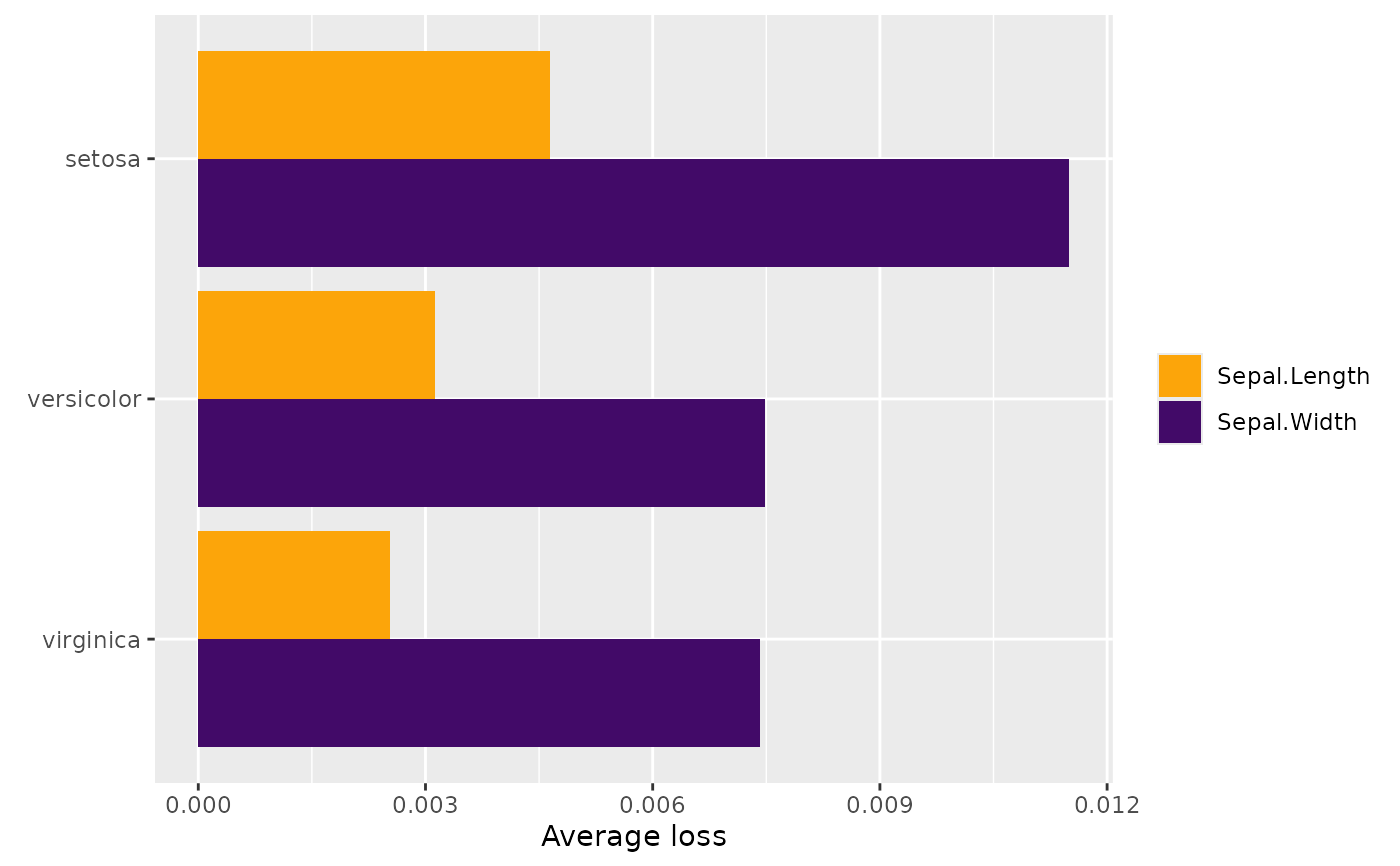
L <- average_loss(
fit, X = iris, y = iris[, 1:2], loss = "gamma", BY = "Species"
)
L
#> Average loss
#> Sepal.Length Sepal.Width
#> setosa 0.004646018 0.011500586
#> versicolor 0.003121888 0.007489254
#> virginica 0.002525590 0.007419552
plot(L)