Print Model Drift Data Frame
Print Model Drift Data Frame
# S3 method for model_drift print(x, max_length = 25, ...)
Arguments
| x | an object of the class `model_drift` |
|---|---|
| max_length | length of the first column, by default 25 |
| ... | other arguments, currently ignored |
Value
this function prints a data frame with a nicer format
Examples
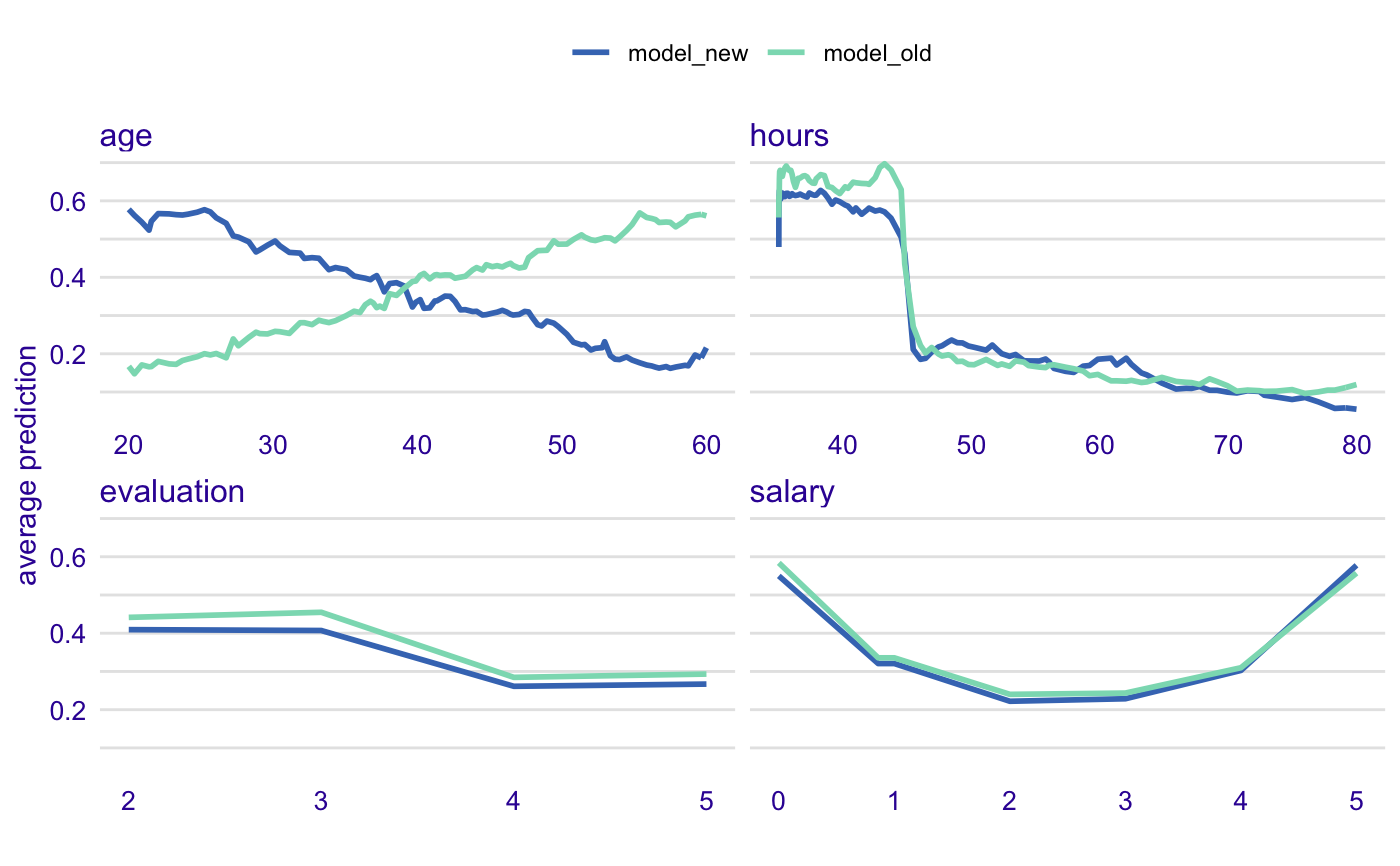
library("DALEX") model_old <- lm(m2.price ~ ., data = apartments) model_new <- lm(m2.price ~ ., data = apartments_test[1:1000,]) calculate_model_drift(model_old, model_new, apartments_test[1:1000,], apartments_test[1:1000,]$m2.price)#> Variable Shift Scaled #> ----------------------------------------------- #> floor 170.48 19.1 . #> no.rooms 168.87 18.9 . #> surface 162.15 18.2 . #> m2.price 169.04 18.9 . #> construction.year 168.95 18.9 .library("ranger") predict_function <- function(m,x,...) predict(m, x, ...)$predictions model_old <- ranger(m2.price ~ ., data = apartments) model_new <- ranger(m2.price ~ ., data = apartments_test) calculate_model_drift(model_old, model_new, apartments_test, apartments_test$m2.price, predict_function = predict_function)#> Variable Shift Scaled #> ----------------------------------------------- #> floor 112.89 12.5 . #> no.rooms 61.11 6.8 #> surface 41.19 4.6 #> m2.price 72.49 8.1 #> construction.year 84.20 9.4# here we compare model created on male data # with model applied to female data # there is interaction with age, and it is detected here predict_function <- function(m,x,...) predict(m, x, ..., probability=TRUE)$predictions[,1] data_old = HR[HR$gender == "male", -1] data_new = HR[HR$gender == "female", -1] model_old <- ranger(status ~ ., data = data_old, probability=TRUE) model_new <- ranger(status ~ ., data = data_new, probability=TRUE) calculate_model_drift(model_old, model_new, HR_test, HR_test$status == "fired", predict_function = predict_function)#> Variable Shift Scaled #> ----------------------------------------------- #> salary 0.11 22.2 * #> evaluation 0.06 13.2 . #> age 0.30 62.2 *** #> hours 0.05 10.4 .# plot it library("ingredients") prof_old <- partial_dependency(model_old, data = data_new[1:1000,], label = "model_old", predict_function = predict_function, grid_points = 101, variable_splits = NULL) prof_new <- partial_dependency(model_new, data = data_new[1:1000,], label = "model_new", predict_function = predict_function, grid_points = 101, variable_splits = NULL) plot(prof_old, prof_new, color = "_label_")