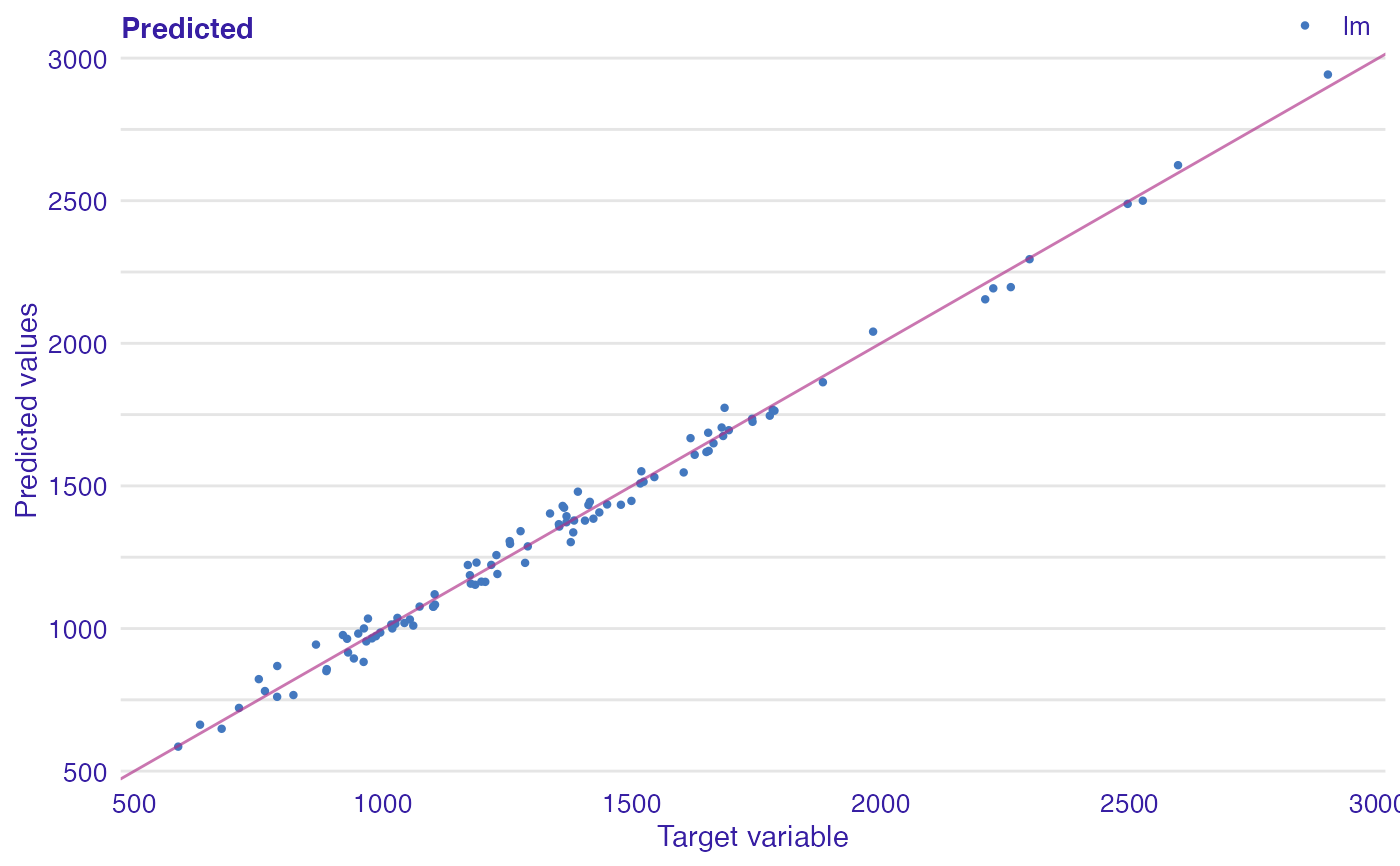
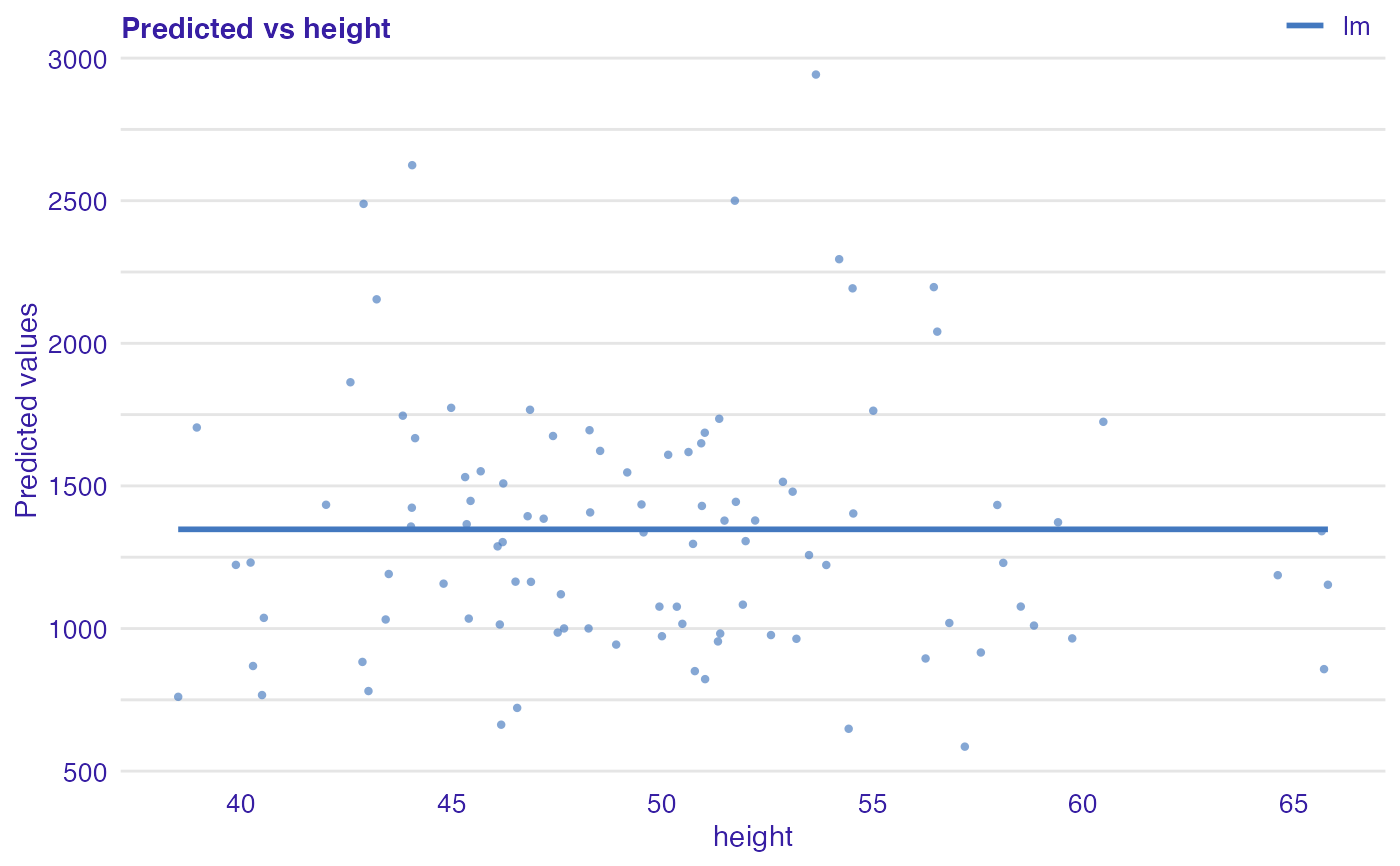
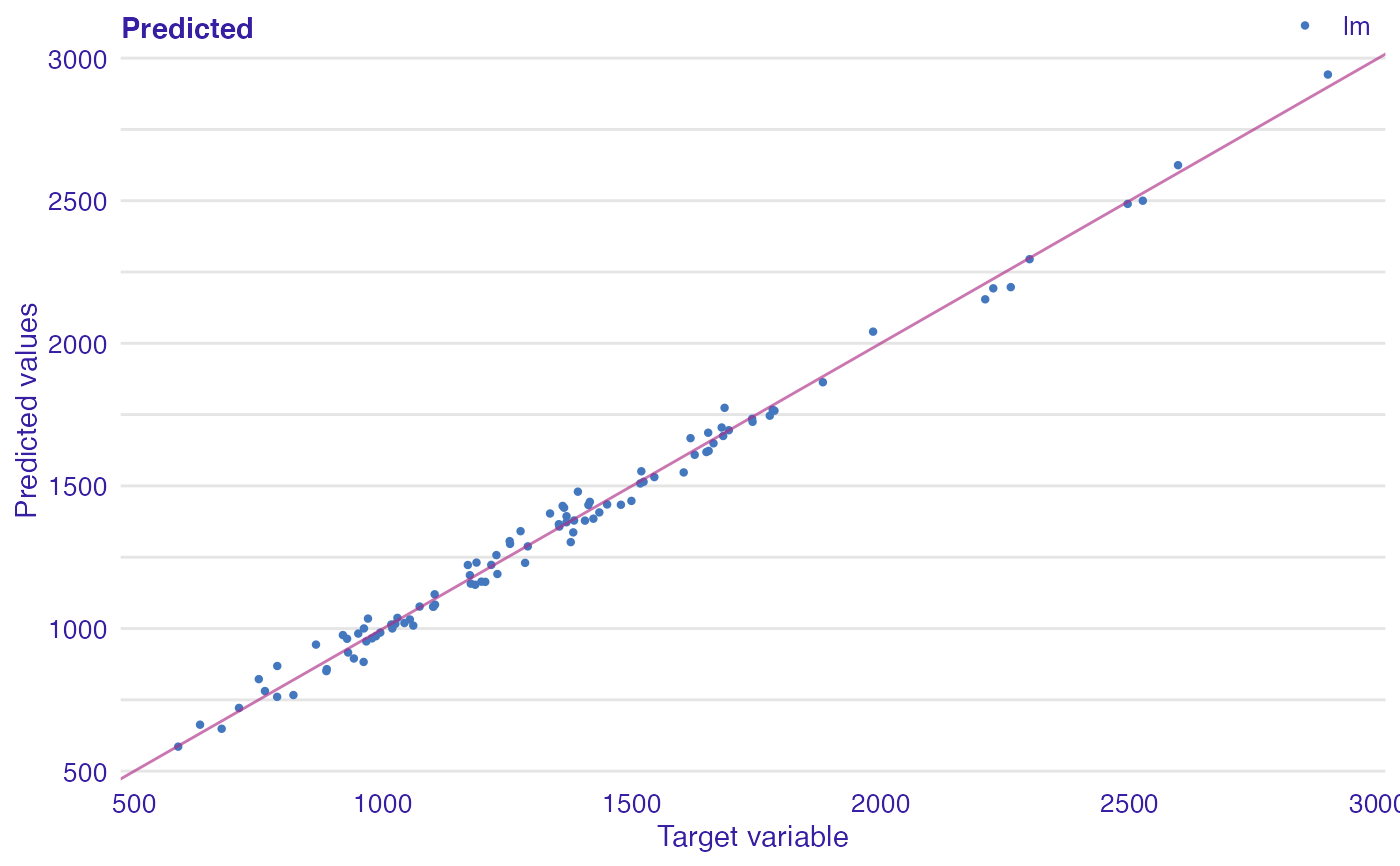
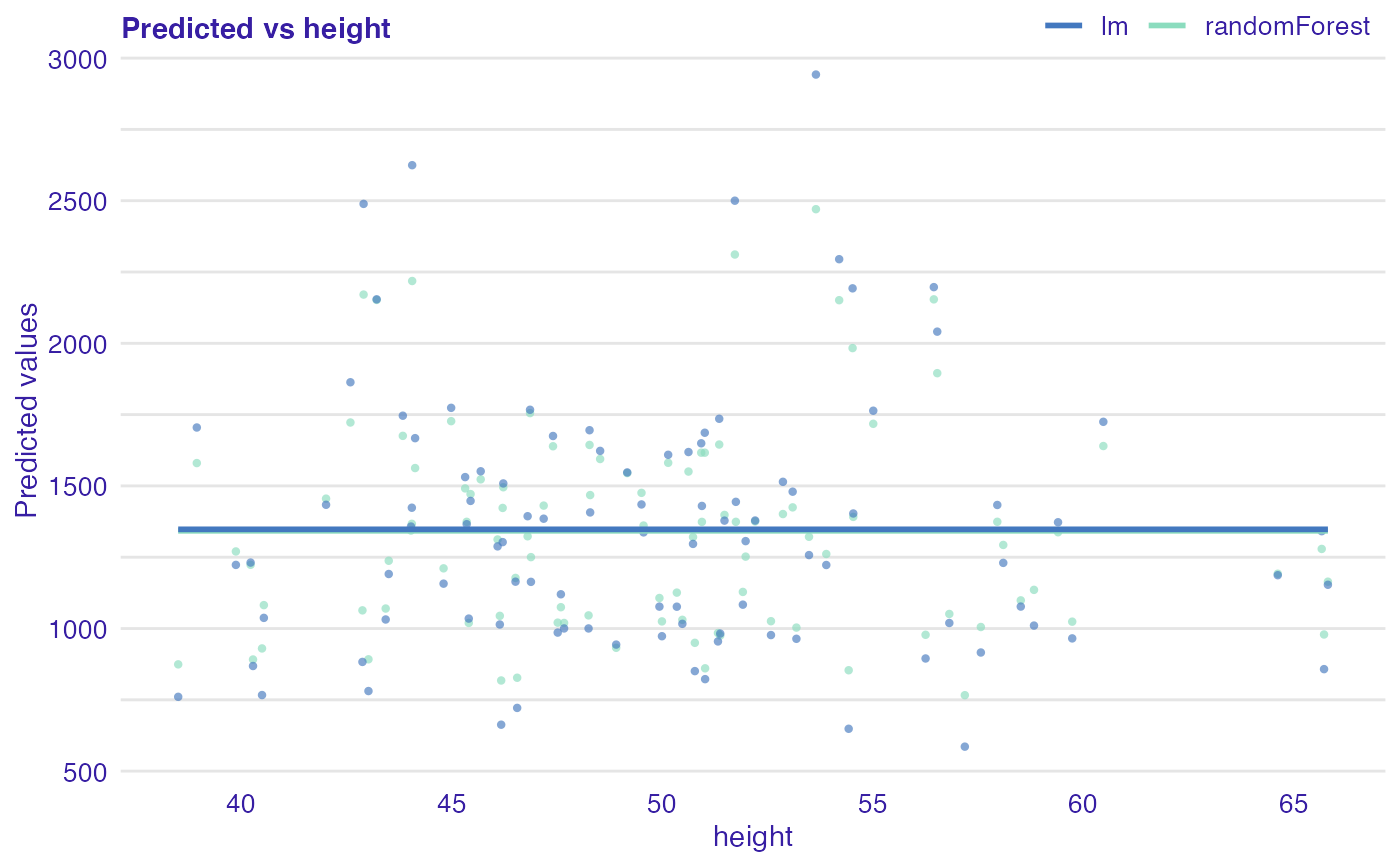
Plot of predicted response vs observed or variable Values.
plot_prediction(object, ..., variable = "_y_", smooth = FALSE, abline = FALSE) plotPrediction(object, ..., variable = NULL, smooth = FALSE, abline = FALSE)
Arguments
| object | An object of class |
|---|---|
| ... | Other |
| variable | Name of variable to order residuals on a plot.
If |
| smooth | Logical, indicates whenever smooth line should be added. |
| abline | Logical, indicates whenever function |
Value
A ggplot2 object.
Examples
dragons <- DALEX::dragons[1:100, ] # fit a model model_lm <- lm(life_length ~ ., data = dragons) lm_audit <- audit(model_lm, data = dragons, y = dragons$life_length)#> Preparation of a new explainer is initiated #> -> model label : lm ( default ) #> -> data : 100 rows 8 cols #> -> target variable : 100 values #> -> predict function : yhat.lm will be used ( default ) #> -> predicted values : No value for predict function target column. ( default ) #> -> model_info : package stats , ver. 4.1.1 , task regression ( default ) #> -> predicted values : numerical, min = 585.8311 , mean = 1347.787 , max = 2942.307 #> -> residual function : difference between y and yhat ( default ) #> -> residuals : numerical, min = -88.41755 , mean = -1.489291e-13 , max = 77.92805 #> A new explainer has been created!# validate a model with auditor mr_lm <- model_residual(lm_audit) # plot results plot_prediction(mr_lm, abline = TRUE)plot_prediction(mr_lm, variable = "height", smooth = TRUE)library(randomForest) model_rf <- randomForest(life_length~., data = dragons) rf_audit <- audit(model_rf, data = dragons, y = dragons$life_length)#> Preparation of a new explainer is initiated #> -> model label : randomForest ( default ) #> -> data : 100 rows 8 cols #> -> target variable : 100 values #> -> predict function : yhat.randomForest will be used ( default ) #> -> predicted values : No value for predict function target column. ( default ) #> -> model_info : package randomForest , ver. 4.6.14 , task regression ( default ) #> -> predicted values : numerical, min = 766.2776 , mean = 1342.534 , max = 2470.308 #> -> residual function : difference between y and yhat ( default ) #> -> residuals : numerical, min = -185.7102 , mean = 5.252424 , max = 428.1124 #> A new explainer has been created!