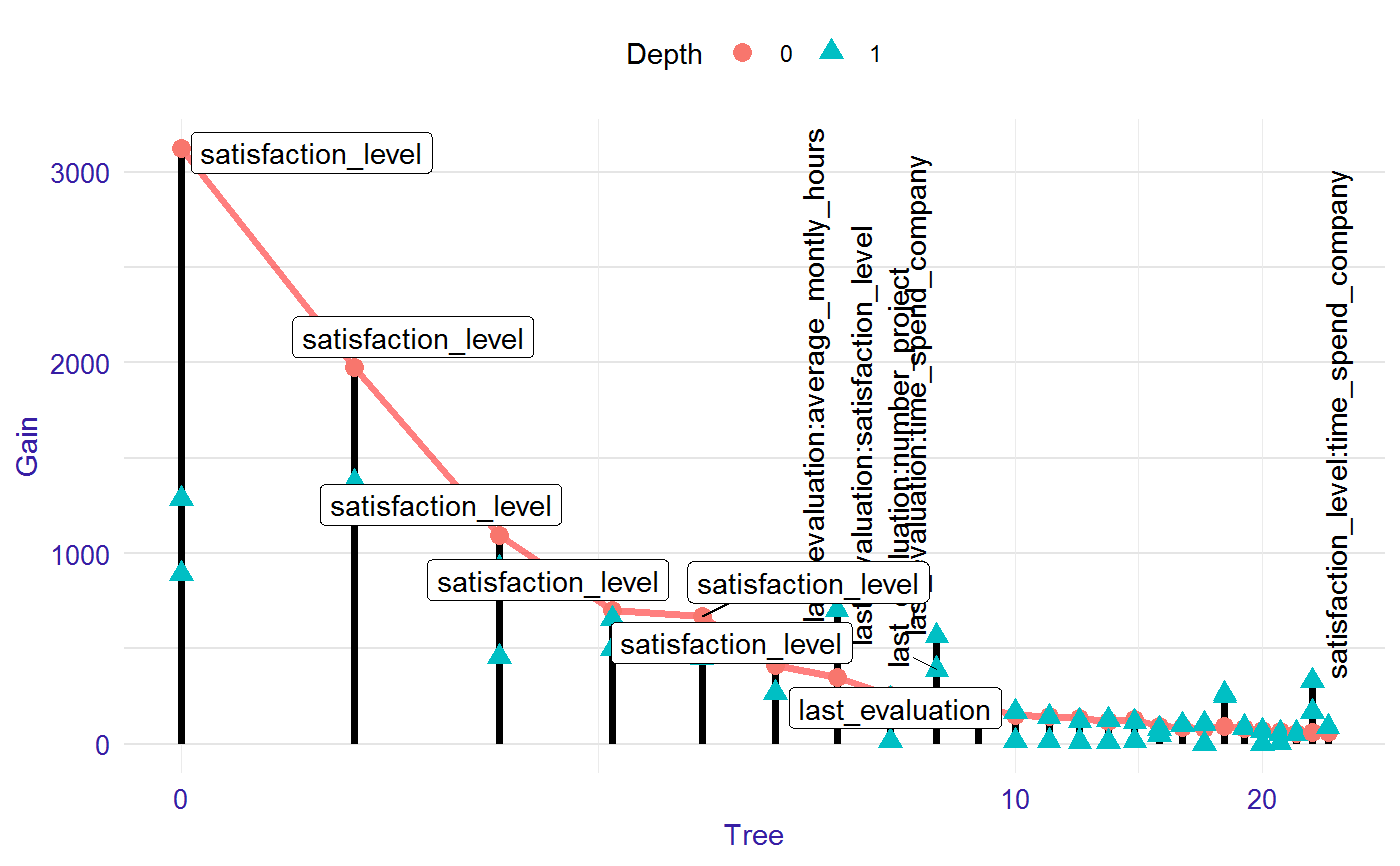
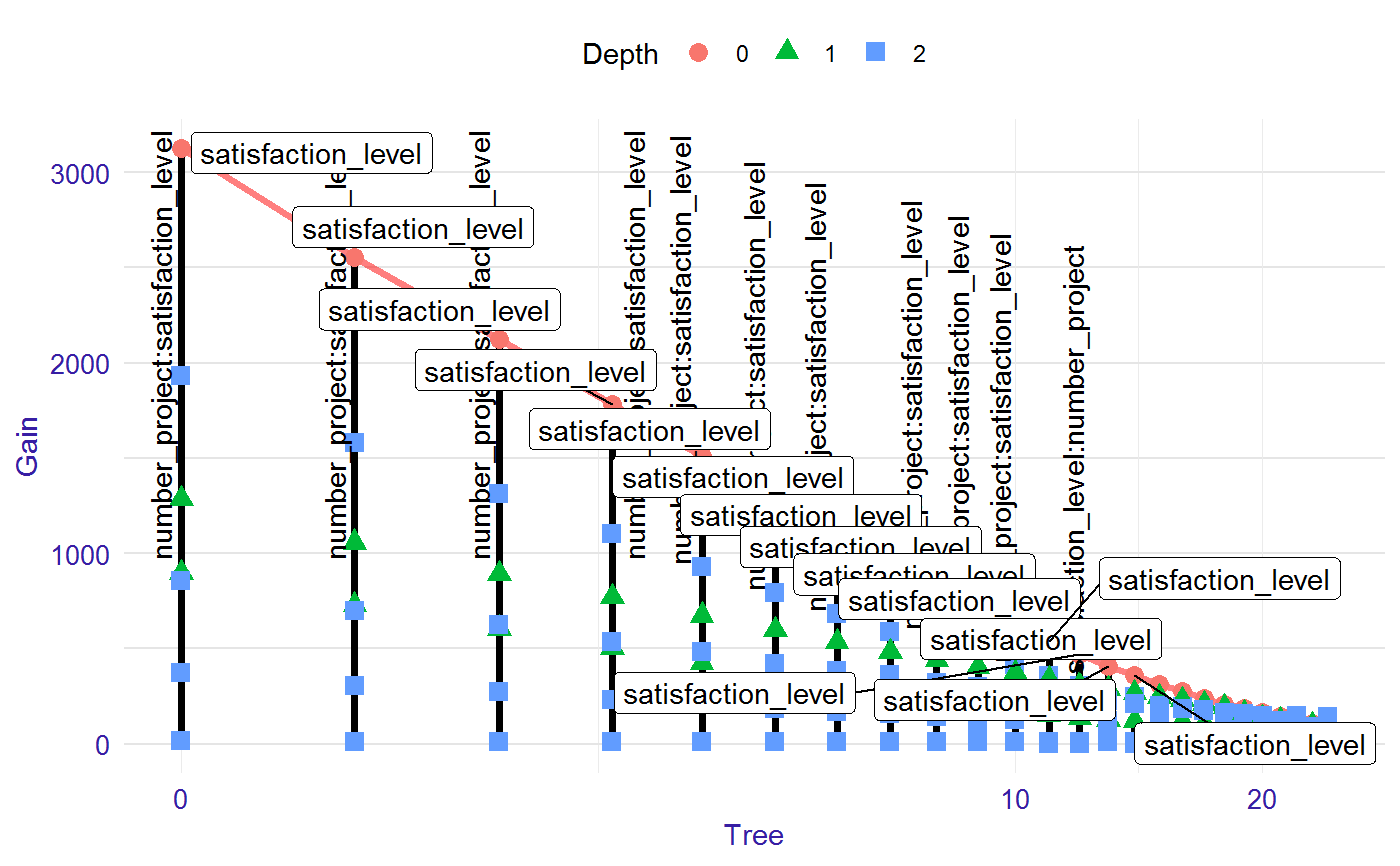
Visualiation of the model
The lollipop plots the model with the most important interactions and variables in the roots.
# S3 method for lollipop plot(x, ..., labels = "topAll", log_scale = TRUE, threshold = 0.1)
Arguments
| x | a result from the |
|---|---|
| ... | other parameters. |
| labels | if "topAll" then labels for the most important interactions (vertical label) and variables in the roots (horizontal label) will be displayed, if "interactions" then labels for all interactions, if "roots" then labels for all variables in the root. |
| log_scale | TRUE/FALSE logarithmic scale on the plot. Default TRUE. |
| threshold | on the plot will occur only labels with Gain higher than `threshold` of the max Gain value in the model. The lower threshold, the more labels on the plot. Range from 0 to 1. Default 0.1. |
Value
a ggplot object
Examples
library("EIX") library("Matrix") sm <- sparse.model.matrix(left ~ . - 1, data = HR_data) library("xgboost") param <- list(objective = "binary:logistic", max_depth = 2) xgb_model <- xgboost(sm, params = param, label = HR_data[, left] == 1, nrounds = 25, verbose = 0) lolli <- lollipop(xgb_model, sm) plot(lolli, labels = "topAll", log_scale = TRUE)library(lightgbm) train_data <- lgb.Dataset(sm, label = HR_data[, left] == 1) params <- list(objective = "binary", max_depth = 3) lgb_model <- lgb.train(params, train_data, 25) lolli <- lollipop(lgb_model, sm) plot(lolli, labels = "topAll", log_scale = TRUE)