Stacked metrics is like plot for chosen_metric but with all unique metrics stacked on top of each other.
Metrics containing NA's will be dropped to enable fair comparison.
# S3 method for stacked_metrics
plot(x, ...)Arguments
| x |
|
|---|---|
| ... | other plot parameters |
Value
ggplot2 object
Examples
data("german")
y_numeric <- as.numeric(german$Risk) - 1
lm_model <- glm(Risk ~ .,
data = german,
family = binomial(link = "logit")
)
explainer_lm <- DALEX::explain(lm_model, data = german[, -1], y = y_numeric)
#> Preparation of a new explainer is initiated
#> -> model label : lm ( default )
#> -> data : 1000 rows 9 cols
#> -> target variable : 1000 values
#> -> predict function : yhat.glm will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package stats , ver. 4.1.1 , task classification ( default )
#> -> predicted values : numerical, min = 0.1369187 , mean = 0.7 , max = 0.9832426
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.9572803 , mean = 1.940006e-17 , max = 0.8283475
#> A new explainer has been created!
fobject <- fairness_check(explainer_lm,
protected = german$Sex,
privileged = "male"
)
#> Creating fairness classification object
#> -> Privileged subgroup : character ( Ok )
#> -> Protected variable : factor ( Ok )
#> -> Cutoff values for explainers : 0.5 ( for all subgroups )
#> -> Fairness objects : 0 objects
#> -> Checking explainers : 1 in total ( compatible )
#> -> Metric calculation : 13/13 metrics calculated for all models
#> Fairness object created succesfully
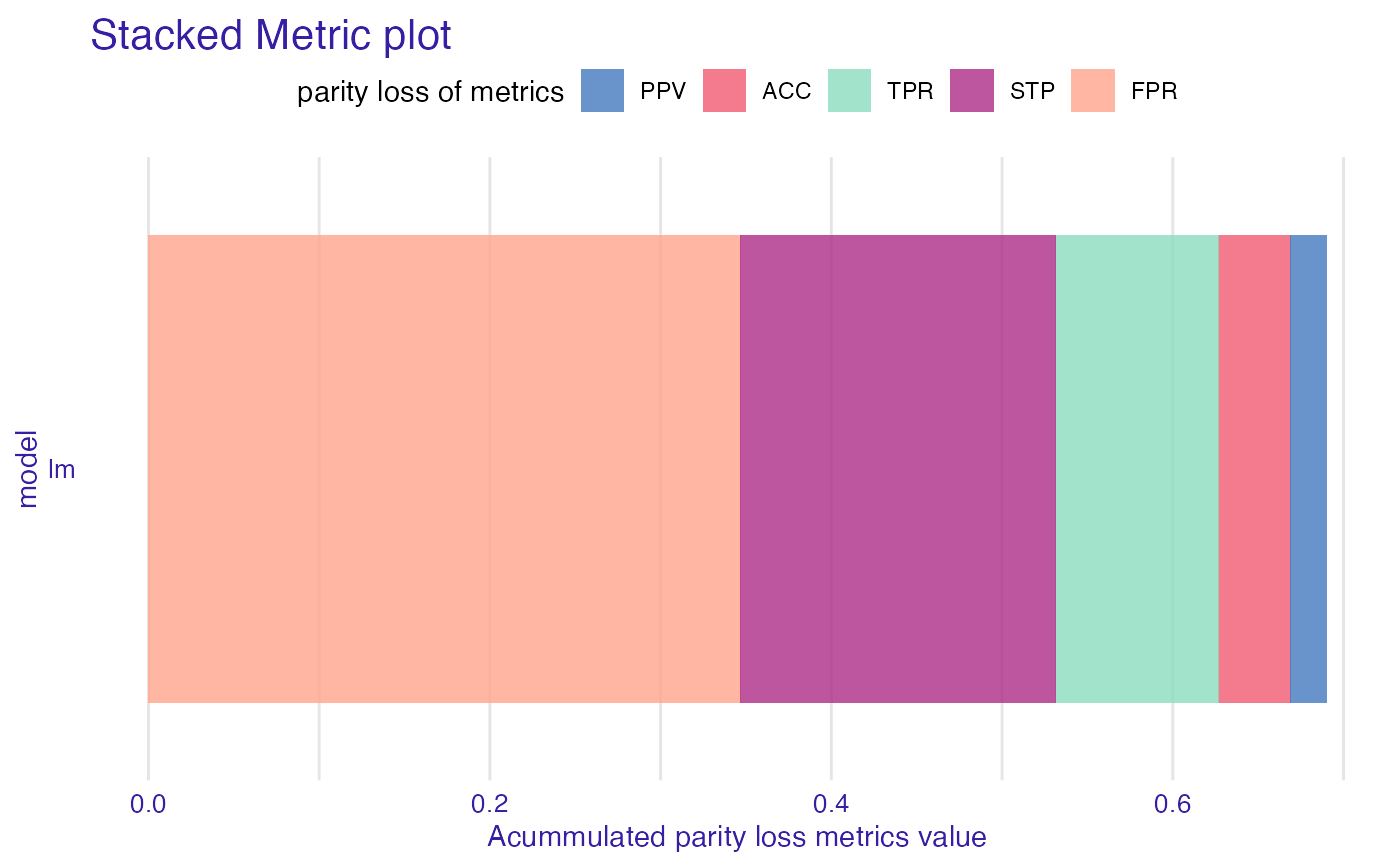
sm <- stack_metrics(fobject)
plot(sm)
 # \donttest{
rf_model <- ranger::ranger(Risk ~ .,
data = german,
probability = TRUE,
num.trees = 200
)
explainer_rf <- DALEX::explain(rf_model, data = german[, -1], y = y_numeric)
#> Preparation of a new explainer is initiated
#> -> model label : ranger ( default )
#> -> data : 1000 rows 9 cols
#> -> target variable : 1000 values
#> -> predict function : yhat.ranger will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package ranger , ver. 0.13.1 , task classification ( default )
#> -> predicted values : numerical, min = 0.07724332 , mean = 0.6974389 , max = 1
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.7611607 , mean = 0.002561123 , max = 0.6776113
#> A new explainer has been created!
fobject <- fairness_check(explainer_rf, fobject)
#> Creating fairness classification object
#> -> Privileged subgroup : character ( from first fairness object )
#> -> Protected variable : factor ( from first fairness object )
#> -> Cutoff values for explainers : 0.5 ( for all subgroups )
#> -> Fairness objects : 1 object ( compatible )
#> -> Checking explainers : 2 in total ( compatible )
#> -> Metric calculation : 13/13 metrics calculated for all models
#> Fairness object created succesfully
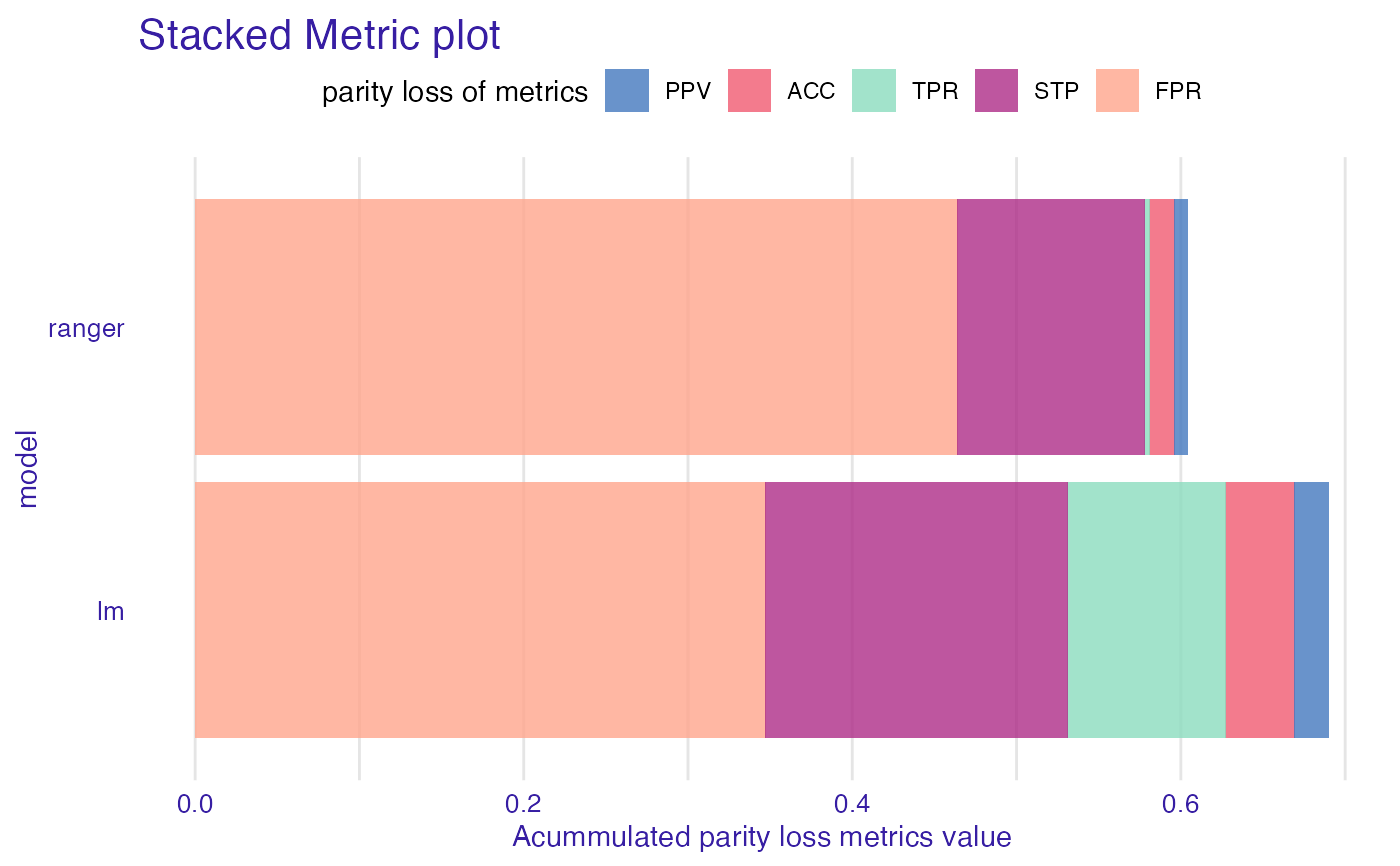
sm <- stack_metrics(fobject)
plot(sm)
# \donttest{
rf_model <- ranger::ranger(Risk ~ .,
data = german,
probability = TRUE,
num.trees = 200
)
explainer_rf <- DALEX::explain(rf_model, data = german[, -1], y = y_numeric)
#> Preparation of a new explainer is initiated
#> -> model label : ranger ( default )
#> -> data : 1000 rows 9 cols
#> -> target variable : 1000 values
#> -> predict function : yhat.ranger will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package ranger , ver. 0.13.1 , task classification ( default )
#> -> predicted values : numerical, min = 0.07724332 , mean = 0.6974389 , max = 1
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.7611607 , mean = 0.002561123 , max = 0.6776113
#> A new explainer has been created!
fobject <- fairness_check(explainer_rf, fobject)
#> Creating fairness classification object
#> -> Privileged subgroup : character ( from first fairness object )
#> -> Protected variable : factor ( from first fairness object )
#> -> Cutoff values for explainers : 0.5 ( for all subgroups )
#> -> Fairness objects : 1 object ( compatible )
#> -> Checking explainers : 2 in total ( compatible )
#> -> Metric calculation : 13/13 metrics calculated for all models
#> Fairness object created succesfully
sm <- stack_metrics(fobject)
plot(sm)
 # }
# }