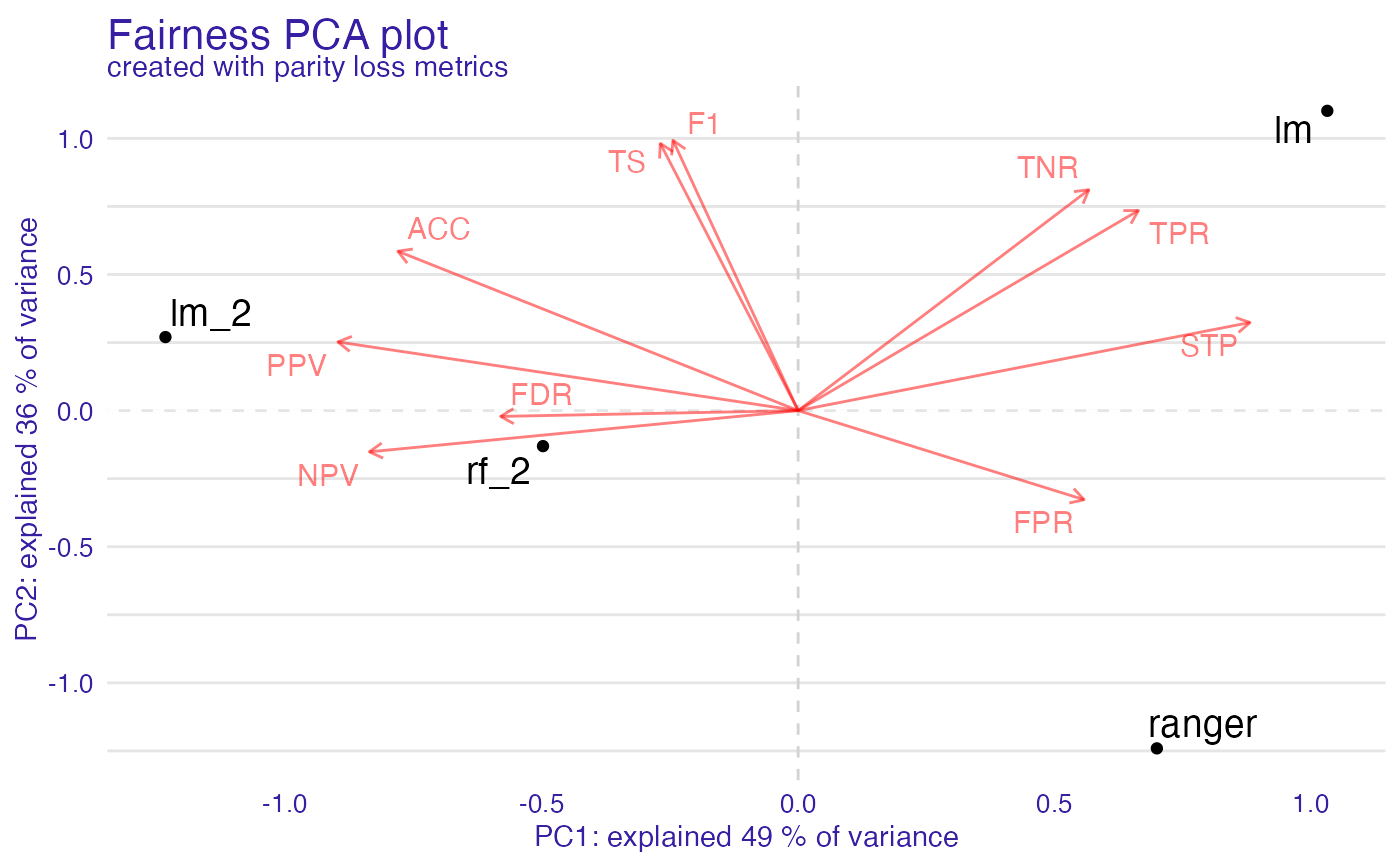
Calculate PC for metric_matrix to see similarities between models and metrics. If omit_models_with_NA is set to TRUE models with NA will be omitted as opposed
to default behavior, when metrics are omitted.
fairness_pca(x, omit_models_with_NA = FALSE)Arguments
| x | object of class |
|---|---|
| omit_models_with_NA | logical, if |
Value
fairness_pca object
It is list containing following fields:
pc_1_2 - amount of data variance explained with each component
rotation - rotation from
stats::prcompx - x from
stats::prcompsdev - sdev from
stats::prcomplabel - model labels
Examples
data("german")
y_numeric <- as.numeric(german$Risk) - 1
lm_model <- glm(Risk ~ .,
data = german,
family = binomial(link = "logit")
)
rf_model <- ranger::ranger(Risk ~ .,
data = german,
probability = TRUE,
num.trees = 200,
num.threads = 1
)
explainer_lm <- DALEX::explain(lm_model, data = german[, -1], y = y_numeric)
#> Preparation of a new explainer is initiated
#> -> model label : lm ( default )
#> -> data : 1000 rows 9 cols
#> -> target variable : 1000 values
#> -> predict function : yhat.glm will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package stats , ver. 4.1.1 , task classification ( default )
#> -> predicted values : numerical, min = 0.1369187 , mean = 0.7 , max = 0.9832426
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.9572803 , mean = 1.940006e-17 , max = 0.8283475
#> A new explainer has been created!
explainer_rf <- DALEX::explain(rf_model, data = german[, -1], y = y_numeric)
#> Preparation of a new explainer is initiated
#> -> model label : ranger ( default )
#> -> data : 1000 rows 9 cols
#> -> target variable : 1000 values
#> -> predict function : yhat.ranger will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package ranger , ver. 0.13.1 , task classification ( default )
#> -> predicted values : numerical, min = 0.05815476 , mean = 0.6969663 , max = 0.9975069
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.7217679 , mean = 0.003033675 , max = 0.6242611
#> A new explainer has been created!
fobject <- fairness_check(explainer_lm, explainer_rf,
protected = german$Sex,
privileged = "male"
)
#> Creating fairness classification object
#> -> Privileged subgroup : character ( Ok )
#> -> Protected variable : factor ( Ok )
#> -> Cutoff values for explainers : 0.5 ( for all subgroups )
#> -> Fairness objects : 0 objects
#> -> Checking explainers : 2 in total ( compatible )
#> -> Metric calculation : 10/13 metrics calculated for all models ( 3 NA created )
#> Fairness object created succesfully
# same explainers with different cutoffs for female
fobject <- fairness_check(explainer_lm, explainer_rf, fobject,
protected = german$Sex,
privileged = "male",
cutoff = list(female = 0.4),
label = c("lm_2", "rf_2")
)
#> Creating fairness classification object
#> -> Privileged subgroup : character ( Ok )
#> -> Protected variable : factor ( Ok )
#> -> Cutoff values for explainers : female: 0.4, male: 0.5
#> -> Fairness objects : 1 object ( compatible )
#> -> Checking explainers : 4 in total ( compatible )
#> -> Metric calculation : 10/13 metrics calculated for all models ( 3 NA created )
#> Fairness object created succesfully
fpca <- fairness_pca(fobject)
#> Warning: Found metric with NA: FNR, FOR, NEW_METRIC, omiting it
plot(fpca)