DALEX is designed to work with various black-box models like tree ensembles, linear models, neural networks etc. Unfortunately R packages that create such models are very inconsistent. Different tools use different interfaces to train, validate and use models. One of those tools, we would like to make more accessible is the xgboost package.
explain_xgboost(
model,
data = NULL,
y = NULL,
weights = NULL,
predict_function = NULL,
predict_function_target_column = NULL,
residual_function = NULL,
...,
label = NULL,
verbose = TRUE,
precalculate = TRUE,
colorize = !isTRUE(getOption("knitr.in.progress")),
model_info = NULL,
type = NULL,
encode_function = NULL,
true_labels = NULL
)Arguments
- model
object - a model to be explained
- data
data.frame or matrix - data which will be used to calculate the explanations. If not provided, then it will be extracted from the model. Data should be passed without a target column (this shall be provided as the
yargument). NOTE: If the target variable is present in thedata, some of the functionalities may not work properly.- y
numeric vector with outputs/scores. If provided, then it shall have the same size as
data- weights
numeric vector with sampling weights. By default it's
NULL. If provided, then it shall have the same length asdata- predict_function
function that takes two arguments: model and new data and returns a numeric vector with predictions. By default it is
yhat.- predict_function_target_column
Character or numeric containing either column name or column number in the model prediction object of the class that should be considered as positive (i.e. the class that is associated with probability 1). If NULL, the second column of the output will be taken for binary classification. For a multiclass classification setting, that parameter cause switch to binary classification mode with one vs others probabilities.
- residual_function
function that takes four arguments: model, data, target vector y and predict function (optionally). It should return a numeric vector with model residuals for given data. If not provided, response residuals (\(y-\hat{y}\)) are calculated. By default it is
residual_function_default.- ...
other parameters
- label
character - the name of the model. By default it's extracted from the 'class' attribute of the model
- verbose
logical. If TRUE (default) then diagnostic messages will be printed
- precalculate
logical. If TRUE (default) then
predicted_valuesandresidualare calculated when explainer is created. This will happen also ifverboseis TRUE. Set bothverboseandprecalculateto FALSE to omit calculations.- colorize
logical. If TRUE (default) then
WARNINGS,ERRORSandNOTESare colorized. Will work only in the R console. Now by default it isFALSEwhile knitting andTRUEotherwise.- model_info
a named list (
package,version,type) containing information about model. IfNULL,DALEXwill seek for information on it's own.- type
type of a model, either
classificationorregression. If not specified thentypewill be extracted frommodel_info.- encode_function
function(data, ...) that if executed with
dataparameters returns encoded dataframe that was used to fit model. Xgboost does not handle factors on it's own so such function is needed to acquire better explanations.- true_labels
a vector of
ybefore encoding.
Value
explainer object (explain) ready to work with DALEX
Examples
library("xgboost")
#>
#> Attaching package: ‘xgboost’
#> The following object is masked from ‘package:dplyr’:
#>
#> slice
library("DALEXtra")
library("mlr")
# 8th column is target that has to be omitted in X data
data <- as.matrix(createDummyFeatures(titanic_imputed[,-8]))
model <- xgboost(data, titanic_imputed$survived, nrounds = 10,
params = list(objective = "binary:logistic"),
prediction = TRUE)
#> [00:35:43] WARNING: amalgamation/../src/learner.cc:627:
#> Parameters: { "prediction" } might not be used.
#>
#> This could be a false alarm, with some parameters getting used by language bindings but
#> then being mistakenly passed down to XGBoost core, or some parameter actually being used
#> but getting flagged wrongly here. Please open an issue if you find any such cases.
#>
#>
#> [1] train-logloss:0.574286
#> [2] train-logloss:0.509887
#> [3] train-logloss:0.472243
#> [4] train-logloss:0.447027
#> [5] train-logloss:0.428637
#> [6] train-logloss:0.415121
#> [7] train-logloss:0.405934
#> [8] train-logloss:0.399623
#> [9] train-logloss:0.394859
#> [10] train-logloss:0.391072
# explainer with encode functiom
explainer_1 <- explain_xgboost(model, data = titanic_imputed[,-8],
titanic_imputed$survived,
encode_function = function(data) {
as.matrix(createDummyFeatures(data))
})
#> Preparation of a new explainer is initiated
#> -> model label : xgb.Booster ( default )
#> -> data : 2207 rows 7 cols
#> -> target variable : 2207 values
#> -> predict function : yhat.xgb.Booster will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package xgboost , ver. 1.6.0.1 , task classification ( default )
#> -> predicted values : numerical, min = 0.04738454 , mean = 0.3272404 , max = 0.9705966
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.8105438 , mean = -0.005083592 , max = 0.9452911
#> A new explainer has been created!
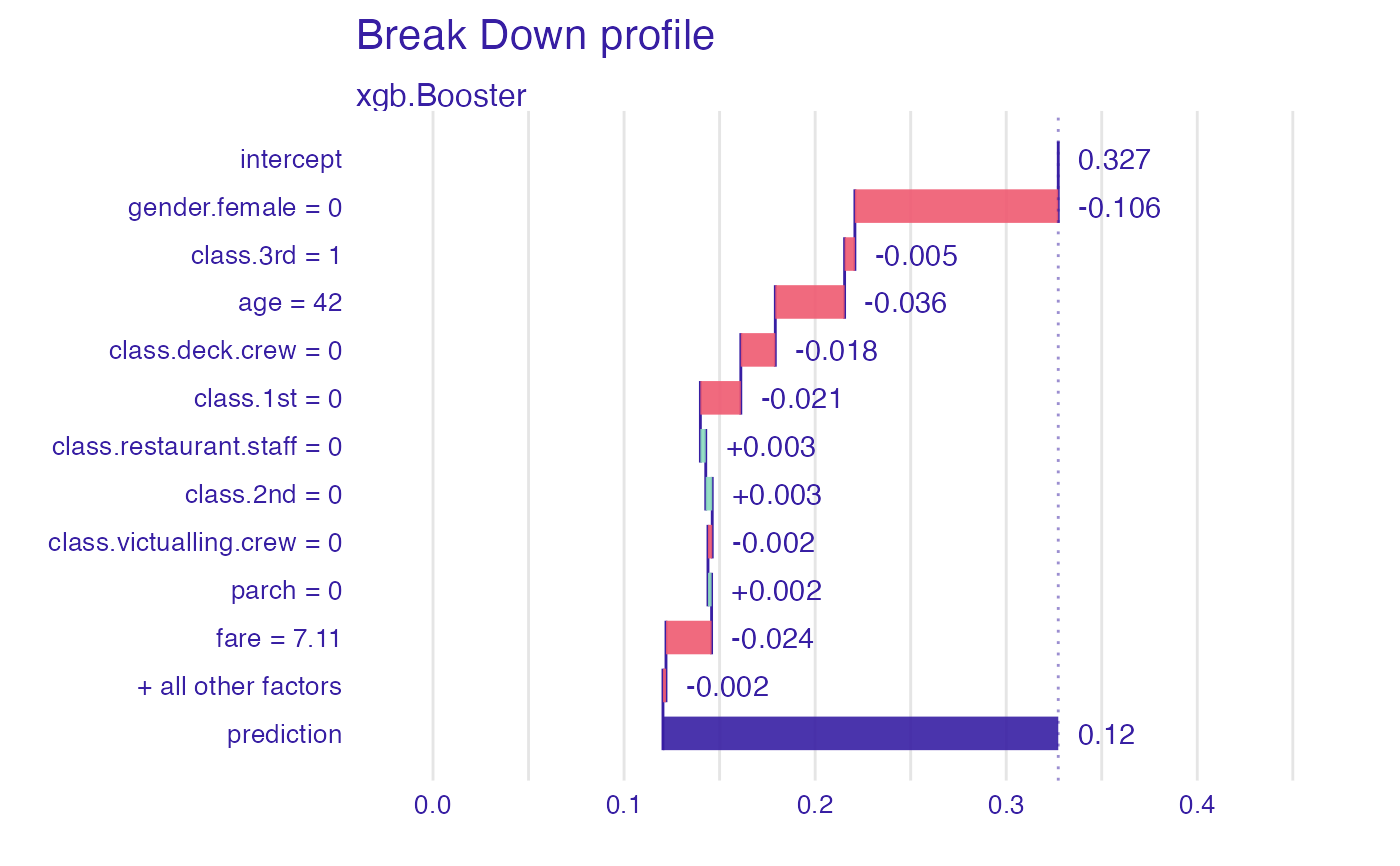
plot(predict_parts(explainer_1, titanic_imputed[1,-8]))
 # explainer without encode function
explainer_2 <- explain_xgboost(model, data = data, titanic_imputed$survived)
#> Preparation of a new explainer is initiated
#> -> model label : xgb.Booster ( default )
#> -> data : 2207 rows 17 cols
#> -> target variable : 2207 values
#> -> predict function : yhat.xgb.Booster will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package xgboost , ver. 1.6.0.1 , task classification ( default )
#> -> predicted values : numerical, min = 0.04738454 , mean = 0.3272404 , max = 0.9705966
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.8105438 , mean = -0.005083592 , max = 0.9452911
#> A new explainer has been created!
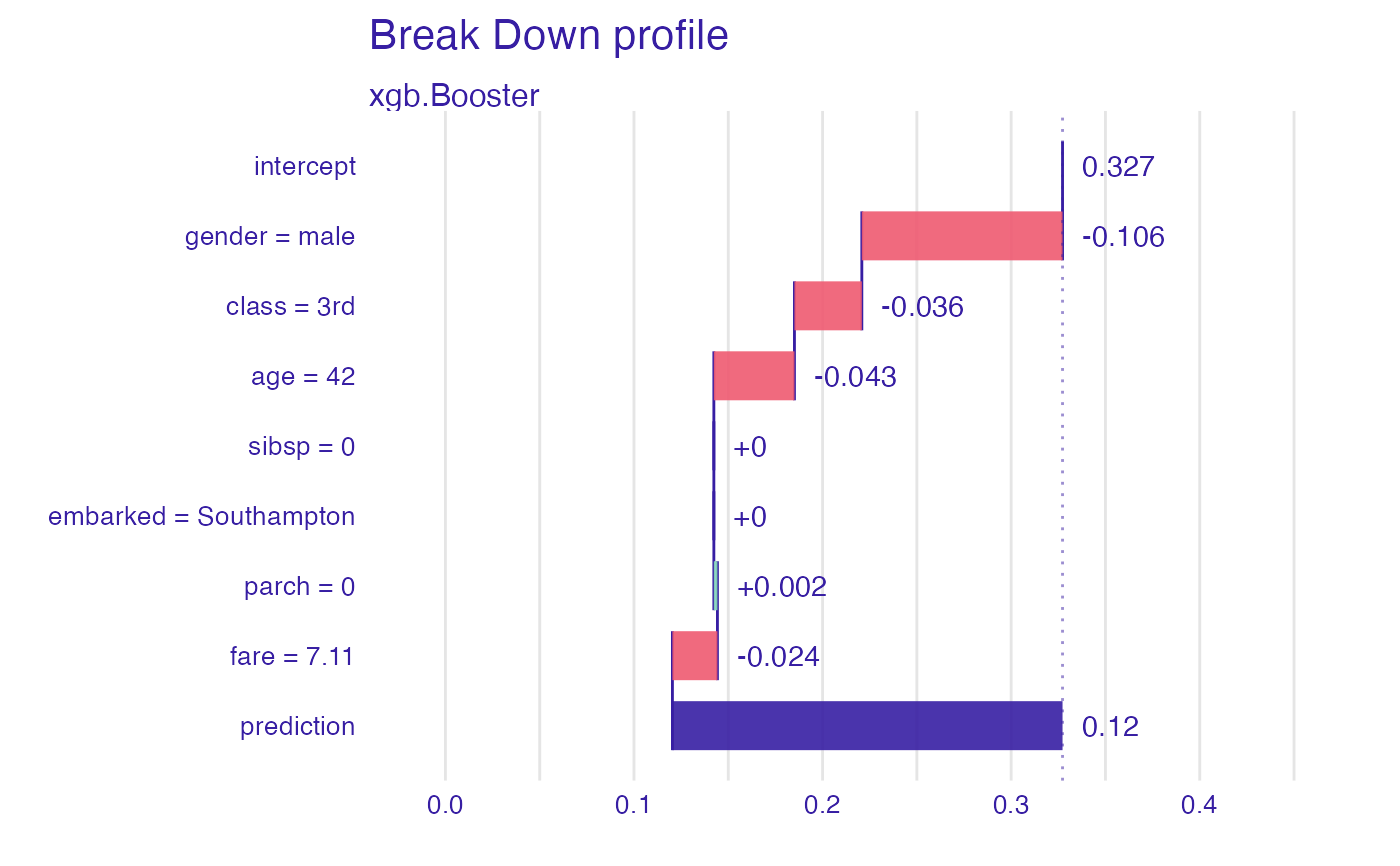
plot(predict_parts(explainer_2, data[1,,drop = FALSE]))
# explainer without encode function
explainer_2 <- explain_xgboost(model, data = data, titanic_imputed$survived)
#> Preparation of a new explainer is initiated
#> -> model label : xgb.Booster ( default )
#> -> data : 2207 rows 17 cols
#> -> target variable : 2207 values
#> -> predict function : yhat.xgb.Booster will be used ( default )
#> -> predicted values : No value for predict function target column. ( default )
#> -> model_info : package xgboost , ver. 1.6.0.1 , task classification ( default )
#> -> predicted values : numerical, min = 0.04738454 , mean = 0.3272404 , max = 0.9705966
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.8105438 , mean = -0.005083592 , max = 0.9452911
#> A new explainer has been created!
plot(predict_parts(explainer_2, data[1,,drop = FALSE]))